bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0016_orf2
Length=121
Score E
Sequences producing significant alignments: (Bits) Value
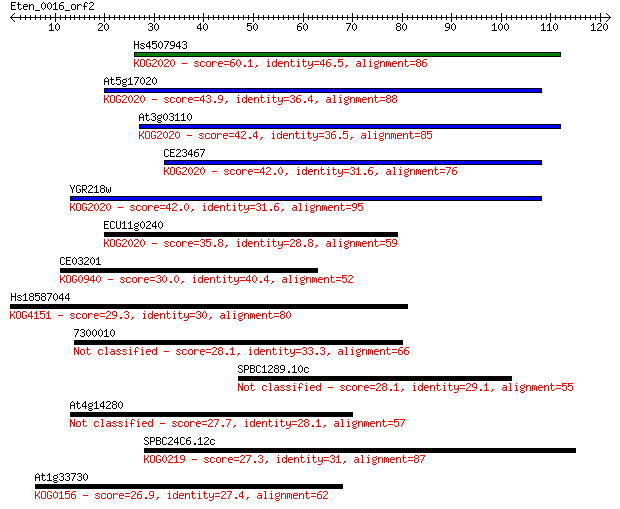
Hs4507943 60.1 1e-09
At5g17020 43.9 8e-05
At3g03110 42.4 2e-04
CE23467 42.0 3e-04
YGR218w 42.0 3e-04
ECU11g0240 35.8 0.022
CE03201 30.0 1.2
Hs18587044 29.3 1.6
7300010 28.1 4.3
SPBC1289.10c 28.1 4.6
At4g14280 27.7 5.8
SPBC24C6.12c 27.3 7.0
At1g33730 26.9 8.8
> Hs4507943
Length=1071
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 40/88 (45%), Positives = 55/88 (62%), Gaps = 5/88 (5%)
Query 26 DLLCRSFQTLNPKQVEAFVLDLFNFCVDSNPSRFQEHMRDFLISLKEFAG-DNEALFEAE 84
+LL +F L QV+ FV LF+ ++ + F+EH+RDFL+ +KEFAG D LF E
Sbjct 980 NLLKSAFPHLQDAQVKLFVTGLFS--LNQDIPAFKEHLRDFLVQIKEFAGEDTSDLFLEE 1037
Query 85 RKEALARAAELEKQKRGM-VPGLLPQYE 111
R+ AL R A+ EK KR M VPG+ +E
Sbjct 1038 REIAL-RQADEEKHKRQMSVPGIFNPHE 1064
> At5g17020
Length=1075
Score = 43.9 bits (102), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 32/89 (35%), Positives = 48/89 (53%), Gaps = 7/89 (7%)
Query 20 VTKSLIDLLCRSFQTLNPKQVEAFVLDLFNFCVDSNPSRFQEHMRDFLISLKEF-AGDNE 78
V + I LL SF + +V FV L+ ++PS F+ ++RDFL+ KEF A DN+
Sbjct 981 VREYTIKLLSSSFPNMTAAEVTQFVNGLYE--SRNDPSGFKNNIRDFLVQSKEFSAQDNK 1038
Query 79 ALFEAERKEALARAAELEKQKRGMVPGLL 107
L+ E A E E+Q+ +PGL+
Sbjct 1039 DLYAEEA----AAQRERERQRMLSIPGLI 1063
> At3g03110
Length=1022
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 31/88 (35%), Positives = 50/88 (56%), Gaps = 11/88 (12%)
Query 27 LLCRSFQTLNPKQVEAFVLDLFNFCVDSNPSRFQEHMRDFLISLKEF-AGDNEALFEAER 85
LL SF + +V FV L+ ++ RF++++RDFLI KEF A DN+ L+ E
Sbjct 935 LLSSSFPNMTTTEVTQFVNGLYE--SRNDVGRFKDNIRDFLIQSKEFSAQDNKDLYAEE- 991
Query 86 KEALARAAELEKQKRGM--VPGLLPQYE 111
AA++E++++ M +PGL+ E
Sbjct 992 -----AAAQMERERQRMLSIPGLIAPSE 1014
> CE23467
Length=1080
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 24/76 (31%), Positives = 42/76 (55%), Gaps = 5/76 (6%)
Query 32 FQTLNPKQVEAFVLDLFNFCVDSNPSRFQEHMRDFLISLKEFAGDNEALFEAERKEALAR 91
F +N Q+ + F+F ++ S + H+RDFLI +KE G++ + E +EA +
Sbjct 999 FDNMNQDQIRIIIKGFFSF--NTEISSMRNHLRDFLIQIKEHNGEDTSDLYLEEREAEIQ 1056
Query 92 AAELEKQKRGMVPGLL 107
A+ Q++ VPG+L
Sbjct 1057 QAQ---QRKRDVPGIL 1069
> YGR218w
Length=1084
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 30/97 (30%), Positives = 52/97 (53%), Gaps = 4/97 (4%)
Query 13 QGLT-QENVTKSLIDLLCRSFQTLNPKQVEAFVLDLFNFCVDSNPSRFQEHMRDFLISLK 71
QG + Q +++ L ++L +F L +Q+ +F+ L D F+ +RDFL+ +K
Sbjct 983 QGTSNQVYLSQYLANMLSNAFPHLTSEQIASFLSALTKQYKDL--VVFKGTLRDFLVQIK 1040
Query 72 EFAGD-NEALFEAERKEALARAAELEKQKRGMVPGLL 107
E GD + LF +++ AL LE++K + GLL
Sbjct 1041 EVGGDPTDYLFAEDKENALMEQNRLEREKAAKIGGLL 1077
> ECU11g0240
Length=1058
Score = 35.8 bits (81), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 17/59 (28%), Positives = 32/59 (54%), Gaps = 2/59 (3%)
Query 20 VTKSLIDLLCRSFQTLNPKQVEAFVLDLFNFCVDSNPSRFQEHMRDFLISLKEFAGDNE 78
+++ ++ L +SF + + V+ F + LF C D F+EH+ DF + + EF D +
Sbjct 985 LSEYIVGLFVKSFPNITQESVKIFSVGLFELCGDDEI--FKEHVEDFRVKVYEFGTDED 1041
> CE03201
Length=457
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 21/55 (38%), Positives = 32/55 (58%), Gaps = 5/55 (9%)
Query 11 EQQGLTQENVTKSL-IDLLCRSFQTLNPKQVEAFVLDL-FNFCVDSNPSR-FQEH 62
+QQG+ Q+N KSL +D + R F T P+ VE + + C DS+ R F++H
Sbjct 187 QQQGMMQQNREKSLSLDPMRRPFMT--PQDVEQLPMPQGWEMCYDSDGVRYFKDH 239
> Hs18587044
Length=1051
Score = 29.3 bits (64), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 24/81 (29%), Positives = 39/81 (48%), Gaps = 6/81 (7%)
Query 1 RIVAFRLVENEQQGLTQENVTKSLIDLLCRSFQTLNPKQVEAFVLDLFNFCVDSNPSRFQ 60
RI + VENE+ L N+ +++ID L + + + EA VLD + +
Sbjct 239 RICSLMAVENEEMSLAVCNLLQAIIDSLSGEDKREHRGKEEALVLD-----TKKDLKQIT 293
Query 61 EHMRDFLISLKEFA-GDNEAL 80
H+ D L+S K G ++AL
Sbjct 294 SHLLDMLVSKKVSGQGRDQAL 314
> 7300010
Length=710
Score = 28.1 bits (61), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 33/69 (47%), Gaps = 7/69 (10%)
Query 14 GLTQENVTKSLIDLLCRSFQTLNPKQVEAFVLDLFNFCVDSN---PSRFQEHMRDFLISL 70
G N+ K+L++LL +T N +QV DL +N P+R ++ L L
Sbjct 532 GAHLTNIQKTLVELLIGECETENVRQVN----DLPRLYRKTNREVPTRCSSYVEQMLRPL 587
Query 71 KEFAGDNEA 79
K FA NE+
Sbjct 588 KAFAQQNES 596
> SPBC1289.10c
Length=743
Score = 28.1 bits (61), Expect = 4.6, Method: Composition-based stats.
Identities = 16/55 (29%), Positives = 31/55 (56%), Gaps = 9/55 (16%)
Query 47 LFNFCVDSNPSRFQEHMRDFLISLKEFAGDNEALFEAERKEALARAAELEKQKRG 101
L+NF ++NPSR +++++ LK L E + K+ L ++ EK+++G
Sbjct 435 LYNFNGNANPSRLNPALKNYMEELK--------LLEQQNKKRLLLVSQ-EKERKG 480
> At4g14280
Length=848
Score = 27.7 bits (60), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 16/57 (28%), Positives = 27/57 (47%), Gaps = 6/57 (10%)
Query 13 QGLTQENVTKSLIDLLCRSFQTLNPKQVEAFVLDLFNFCVDSNPSRFQEHMRDFLIS 69
Q + + NV K L++ L LN ++ L N C + P +F E M++ + S
Sbjct 604 QKILRANVLKGLVEALDNPLIRLNAARI------LRNLCAYTAPGQFNEQMKEVIKS 654
> SPBC24C6.12c
Length=434
Score = 27.3 bits (59), Expect = 7.0, Method: Composition-based stats.
Identities = 27/100 (27%), Positives = 41/100 (41%), Gaps = 13/100 (13%)
Query 28 LCRSFQTLNPKQVEAFVLD--------LFNFCVDSNPSRFQEH---MRDFLISLKEFAGD 76
L T+ V A+V D L+N C ++ F H + F + E A +
Sbjct 294 LSEEITTVKNLHVTAYVGDSESKDVALLYNVCEGASDRSFGIHVAKLAHFPPKIIEMASN 353
Query 77 NEALFEAERKEALARAAELEKQKRGM--VPGLLPQYESMV 114
A EAE A E++ +K GM V ++ Q+ S V
Sbjct 354 KAAELEAEDSGAQGDTQEVKSKKEGMAIVRDIMRQWRSNV 393
> At1g33730
Length=368
Score = 26.9 bits (58), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 17/66 (25%), Positives = 34/66 (51%), Gaps = 4/66 (6%)
Query 6 RLVEN----EQQGLTQENVTKSLIDLLCRSFQTLNPKQVEAFVLDLFNFCVDSNPSRFQE 61
R++EN +++ ++ + +LIDL +N ++E +LD+F D+N S +
Sbjct 127 RILENSSRIDEKDVSSRDFLDALIDLQQGDESEINIDEIEHLLLDMFLAGTDTNSSTVEW 186
Query 62 HMRDFL 67
M + L
Sbjct 187 AMTELL 192
Lambda K H
0.320 0.135 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1198419392
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40