bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0014_orf1
Length=154
Score E
Sequences producing significant alignments: (Bits) Value
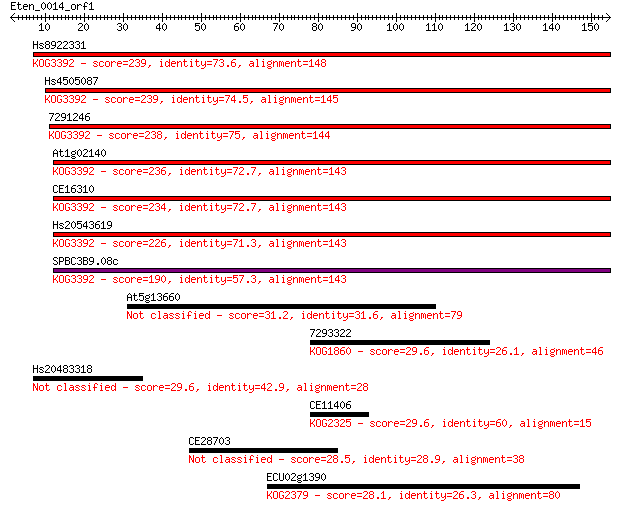
Hs8922331 239 1e-63
Hs4505087 239 2e-63
7291246 238 2e-63
At1g02140 236 1e-62
CE16310 234 7e-62
Hs20543619 226 9e-60
SPBC3B9.08c 190 9e-49
At5g13660 31.2 0.90
7293322 29.6 2.2
Hs20483318 29.6 2.3
CE11406 29.6 2.5
CE28703 28.5 5.3
ECU02g1390 28.1 6.9
> Hs8922331
Length=148
Score = 239 bits (611), Expect = 1e-63, Method: Compositional matrix adjust.
Identities = 109/148 (73%), Positives = 129/148 (87%), Gaps = 0/148 (0%)
Query 7 MGDDDDFYLRYYTGHKGKFGHEFLEFEFLAEGRMRYANNSNYKNDTMIRKEAYVSQAVIK 66
M DFYLRYY GHKGKFGHEFLEFEF +G++RYANNSNYKND MIRKEAYV ++V++
Sbjct 1 MAVASDFYLRYYVGHKGKFGHEFLEFEFRPDGKLRYANNSNYKNDVMIRKEAYVHKSVME 60
Query 67 ELRRIIEEAEILREDDNNWPAPDRVGRQELEIILGKDHISFTTSKIGSMADVQRCKDQEG 126
EL+RII+++EI +EDD WP PDRVGRQELEI++G +HISFTTSKIGS+ DV + KD EG
Sbjct 61 ELKRIIDDSEITKEDDALWPPPDRVGRQELEIVIGDEHISFTTSKIGSLIDVNQSKDPEG 120
Query 127 LRVFYYLVQDLKCFVYSLIGLHFRIKPV 154
LRVFYYLVQDLKC V+SLIGLHF+IKP+
Sbjct 121 LRVFYYLVQDLKCLVFSLIGLHFKIKPI 148
> Hs4505087
Length=146
Score = 239 bits (609), Expect = 2e-63, Method: Compositional matrix adjust.
Identities = 108/145 (74%), Positives = 129/145 (88%), Gaps = 0/145 (0%)
Query 10 DDDFYLRYYTGHKGKFGHEFLEFEFLAEGRMRYANNSNYKNDTMIRKEAYVSQAVIKELR 69
+ DFYLRYY GHKGKFGHEFLEFEF +G++RYANNSNYKND MIRKEAYV ++V++EL+
Sbjct 2 ESDFYLRYYVGHKGKFGHEFLEFEFRPDGKLRYANNSNYKNDVMIRKEAYVHKSVMEELK 61
Query 70 RIIEEAEILREDDNNWPAPDRVGRQELEIILGKDHISFTTSKIGSMADVQRCKDQEGLRV 129
RII+++EI +EDD WP PDRVGRQELEI++G +HISFTTSKIGS+ DV + KD EGLRV
Sbjct 62 RIIDDSEITKEDDALWPPPDRVGRQELEIVIGDEHISFTTSKIGSLIDVNQSKDPEGLRV 121
Query 130 FYYLVQDLKCFVYSLIGLHFRIKPV 154
FYYLVQDLKC V+SLIGLHF+IKP+
Sbjct 122 FYYLVQDLKCLVFSLIGLHFKIKPI 146
> 7291246
Length=147
Score = 238 bits (608), Expect = 2e-63, Method: Compositional matrix adjust.
Identities = 108/144 (75%), Positives = 128/144 (88%), Gaps = 0/144 (0%)
Query 11 DDFYLRYYTGHKGKFGHEFLEFEFLAEGRMRYANNSNYKNDTMIRKEAYVSQAVIKELRR 70
+DFYLRYY GHKGKFGHEFLEFEF +G++RYANNSNYKNDTMIRKEA+V Q+V++EL+R
Sbjct 4 EDFYLRYYVGHKGKFGHEFLEFEFRPDGKLRYANNSNYKNDTMIRKEAFVHQSVMEELKR 63
Query 71 IIEEAEILREDDNNWPAPDRVGRQELEIILGKDHISFTTSKIGSMADVQRCKDQEGLRVF 130
II ++EI++EDD WP PDRVGRQELEI++G +HISFTTSK GS+ DV R KD EGLR F
Sbjct 64 IIIDSEIMQEDDLPWPPPDRVGRQELEIVIGDEHISFTTSKTGSLVDVNRSKDPEGLRCF 123
Query 131 YYLVQDLKCFVYSLIGLHFRIKPV 154
YYLVQDLKC V+SLIGLHF+IKP+
Sbjct 124 YYLVQDLKCLVFSLIGLHFKIKPI 147
> At1g02140
Length=150
Score = 236 bits (602), Expect = 1e-62, Method: Compositional matrix adjust.
Identities = 104/143 (72%), Positives = 125/143 (87%), Gaps = 0/143 (0%)
Query 12 DFYLRYYTGHKGKFGHEFLEFEFLAEGRMRYANNSNYKNDTMIRKEAYVSQAVIKELRRI 71
+FYLRYY GHKGKFGHEFLEFEF +G++RYANNSNYKNDT+IRKE +++ AV+KE +RI
Sbjct 8 EFYLRYYVGHKGKFGHEFLEFEFREDGKLRYANNSNYKNDTIIRKEVFLTPAVLKECKRI 67
Query 72 IEEAEILREDDNNWPAPDRVGRQELEIILGKDHISFTTSKIGSMADVQRCKDQEGLRVFY 131
+ E+EIL+EDDNNWP PDRVG+QELEI+LG +HISF TSKIGS+ D Q D EGLR+FY
Sbjct 68 VSESEILKEDDNNWPEPDRVGKQELEIVLGNEHISFATSKIGSLVDCQSSNDPEGLRIFY 127
Query 132 YLVQDLKCFVYSLIGLHFRIKPV 154
YLVQDLKC V+SLI LHF+IKP+
Sbjct 128 YLVQDLKCLVFSLISLHFKIKPI 150
> CE16310
Length=152
Score = 234 bits (596), Expect = 7e-62, Method: Compositional matrix adjust.
Identities = 104/143 (72%), Positives = 125/143 (87%), Gaps = 0/143 (0%)
Query 12 DFYLRYYTGHKGKFGHEFLEFEFLAEGRMRYANNSNYKNDTMIRKEAYVSQAVIKELRRI 71
DFY+RYY GHKGKFGHEFLEFEF G +RYANNSNYKNDTMIRKEA VS++V+ EL+RI
Sbjct 10 DFYVRYYVGHKGKFGHEFLEFEFRPNGSLRYANNSNYKNDTMIRKEATVSESVLSELKRI 69
Query 72 IEEAEILREDDNNWPAPDRVGRQELEIILGKDHISFTTSKIGSMADVQRCKDQEGLRVFY 131
IE++EI++EDD+NWP PD++GRQELEI+ +HISFTT KIG++ADV KD +GLR FY
Sbjct 70 IEDSEIMQEDDDNWPEPDKIGRQELEILYKNEHISFTTGKIGALADVNNSKDPDGLRSFY 129
Query 132 YLVQDLKCFVYSLIGLHFRIKPV 154
YLVQDLKC V+SLIGLHF+IKP+
Sbjct 130 YLVQDLKCLVFSLIGLHFKIKPI 152
> Hs20543619
Length=148
Score = 226 bits (577), Expect = 9e-60, Method: Compositional matrix adjust.
Identities = 102/143 (71%), Positives = 123/143 (86%), Gaps = 0/143 (0%)
Query 12 DFYLRYYTGHKGKFGHEFLEFEFLAEGRMRYANNSNYKNDTMIRKEAYVSQAVIKELRRI 71
DFYLRYY GHKGKFGHEFLE EF +G++RYAN+SNYKND MIRKEAYV +++++EL+RI
Sbjct 6 DFYLRYYVGHKGKFGHEFLESEFQLDGKLRYANSSNYKNDVMIRKEAYVHKSLMEELKRI 65
Query 72 IEEAEILREDDNNWPAPDRVGRQELEIILGKDHISFTTSKIGSMADVQRCKDQEGLRVFY 131
I++ EI +EDD WP PDR GRQELEI++G +HISFTTSKIGS+ DV + KD EGLRVFY
Sbjct 66 IDDNEITKEDDGLWPPPDRAGRQELEIVIGDEHISFTTSKIGSLIDVNQSKDPEGLRVFY 125
Query 132 YLVQDLKCFVYSLIGLHFRIKPV 154
YLVQDL C V+SLIGLHF+I P+
Sbjct 126 YLVQDLNCLVFSLIGLHFKINPI 148
> SPBC3B9.08c
Length=147
Score = 190 bits (482), Expect = 9e-49, Method: Compositional matrix adjust.
Identities = 82/143 (57%), Positives = 117/143 (81%), Gaps = 0/143 (0%)
Query 12 DFYLRYYTGHKGKFGHEFLEFEFLAEGRMRYANNSNYKNDTMIRKEAYVSQAVIKELRRI 71
DFY+RYY+GH G+FGHEFLEF++ ++G RYANNSNY+ND++IRKE +VS+ V+KE++RI
Sbjct 3 DFYVRYYSGHHGRFGHEFLEFDYHSDGLARYANNSNYRNDSLIRKEMFVSELVLKEVQRI 62
Query 72 IEEAEILREDDNNWPAPDRVGRQELEIILGKDHISFTTSKIGSMADVQRCKDQEGLRVFY 131
++++EI++E D +WP ++ G+QELEI + HI F T K+GS+ADVQ D EGL+VFY
Sbjct 63 VDDSEIIKESDESWPPENKDGKQELEIRMNGKHIMFETCKLGSLADVQNSDDPEGLKVFY 122
Query 132 YLVQDLKCFVYSLIGLHFRIKPV 154
YL+QDLK +SLI L+F+++PV
Sbjct 123 YLIQDLKALCFSLISLNFKLRPV 145
> At5g13660
Length=536
Score = 31.2 bits (69), Expect = 0.90, Method: Composition-based stats.
Identities = 25/90 (27%), Positives = 41/90 (45%), Gaps = 17/90 (18%)
Query 31 EFEFLAEGRMRYANNSNYKNDTMIRKEAYVSQAVIKELRRIIEE-----------AEILR 79
EFE A + ++ NN Y + K Y V++EL+ +E ++I
Sbjct 453 EFEQHANCKTKHPNNHIYFENG---KTIY---GVVQELKNTPQEKLFDAIQNVTGSDINH 506
Query 80 EDDNNWPAPDRVGRQELEIILGKDHISFTT 109
++ N W A V R EL+ I GKD ++ +
Sbjct 507 KNFNTWKASYHVARLELQRIYGKDDVTLAS 536
> 7293322
Length=2122
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 12/46 (26%), Positives = 22/46 (47%), Gaps = 0/46 (0%)
Query 78 LREDDNNWPAPDRVGRQELEIILGKDHISFTTSKIGSMADVQRCKD 123
+R+DD++W A D + ++G D I ++ + RC D
Sbjct 960 IRDDDHDWKATDLAEANGIICLIGLDDIRLLPDRLKPLLQASRCHD 1005
> Hs20483318
Length=311
Score = 29.6 bits (65), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 12/28 (42%), Positives = 16/28 (57%), Gaps = 0/28 (0%)
Query 7 MGDDDDFYLRYYTGHKGKFGHEFLEFEF 34
+GDDD +R+ GHK GH+ E F
Sbjct 16 LGDDDTAEVRWADGHKSVIGHQSQEHVF 43
> CE11406
Length=477
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 9/15 (60%), Positives = 12/15 (80%), Gaps = 0/15 (0%)
Query 78 LREDDNNWPAPDRVG 92
L+EDD NWP PD++
Sbjct 233 LQEDDENWPRPDKLA 247
> CE28703
Length=182
Score = 28.5 bits (62), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 11/38 (28%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 47 NYKNDTMIRKEAYVSQAVIKELRRIIEEAEILREDDNN 84
+Y DT IR + S A+ +I++++ ++ DD+N
Sbjct 32 DYDTDTKIRMLVFTSSAITDSFLKILQQSAVVEVDDSN 69
> ECU02g1390
Length=487
Score = 28.1 bits (61), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 21/80 (26%), Positives = 34/80 (42%), Gaps = 2/80 (2%)
Query 67 ELRRIIEEAEILREDDNNWPAPDRVGRQELEIILGKDHISFTTSKIGSMADVQRCKDQEG 126
ELR++ E +L D W +RV +E G D +S G + + G
Sbjct 251 ELRQVKHETRVLEAGDFLWVRGERVCSSIIERKRGSDFVSSIMD--GRFKEQKNRLRNSG 308
Query 127 LRVFYYLVQDLKCFVYSLIG 146
+R +Y+V+ LK +G
Sbjct 309 IRRIFYVVEGLKNIHMQKVG 328
Lambda K H
0.324 0.142 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1997677330
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40