bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
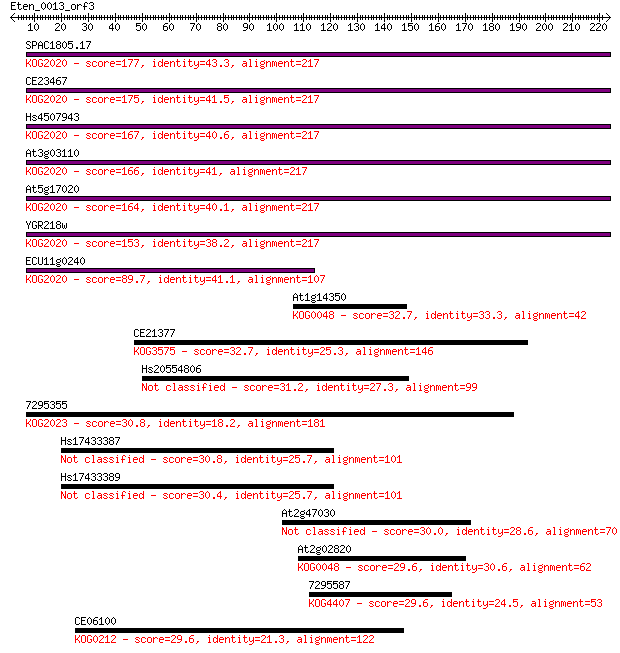
Query= Eten_0013_orf3
Length=223
Score E
Sequences producing significant alignments: (Bits) Value
SPAC1805.17 177 2e-44
CE23467 175 6e-44
Hs4507943 167 2e-41
At3g03110 166 4e-41
At5g17020 164 1e-40
YGR218w 153 3e-37
ECU11g0240 89.7 4e-18
At1g14350 32.7 0.55
CE21377 32.7 0.63
Hs20554806 31.2 1.7
7295355 30.8 2.4
Hs17433387 30.8 2.5
Hs17433389 30.4 3.1
At2g47030 30.0 3.7
At2g02820 29.6 4.9
7295587 29.6 5.5
CE06100 29.6 5.7
> SPAC1805.17
Length=966
Score = 177 bits (448), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 94/218 (43%), Positives = 131/218 (60%), Gaps = 1/218 (0%)
Query 7 VVASNIMYVVGQYPRFLRAYWKFLKTVISKLFEFMQEPFPGVRDMACETFLKVANKCKTT 66
VVASNIMYVVGQYPRFL+A+WKFLKTV++KLFEFM E GV+DMAC+TF+K+A KC+
Sbjct 540 VVASNIMYVVGQYPRFLKAHWKFLKTVVNKLFEFMHEYHEGVQDMACDTFIKIAQKCRRH 599
Query 67 IAANHQEDERNFVLSVIENHTQQTEVLDEKQQLLFFEAVGHIISATPAE-VQASCIEGLM 125
A + F+ +I N + TE L +Q F+EA G++ISA P + +Q I LM
Sbjct 600 FVAQQLGETEPFINEIIRNLAKTTEDLTPQQTHTFYEACGYMISAQPQKHLQERLIFDLM 659
Query 126 ANSWEAWQQIVKSGKENQLFLFNPQAAKRLVQILRINQRVAKSTGTAFTPQLSRLYPEMM 185
A +AW+ IV +N L +PQ K L +L+ N S G+ F PQ+++ Y +M+
Sbjct 660 ALPNQAWENIVAQAAQNAQVLGDPQTVKILANVLKTNVAACTSIGSGFYPQIAKNYVDML 719
Query 186 EIYGVYGQRILQEVAAGGPGRVRHSEIKNLIIFKRETL 223
+Y I + VAA G + ++ L K+E L
Sbjct 720 GLYKAVSGLISEVVAAQGNIATKTPHVRGLRTIKKEIL 757
> CE23467
Length=1080
Score = 175 bits (444), Expect = 6e-44, Method: Compositional matrix adjust.
Identities = 90/218 (41%), Positives = 128/218 (58%), Gaps = 1/218 (0%)
Query 7 VVASNIMYVVGQYPRFLRAYWKFLKTVISKLFEFMQEPFPGVRDMACETFLKVANKCKTT 66
V+ASNIMYVVGQYPRFLRA+WKFLKTVI+KLFEFM E GV+DMAC+TF+K++ KCK
Sbjct 549 VIASNIMYVVGQYPRFLRAHWKFLKTVINKLFEFMHETHEGVQDMACDTFIKISIKCKRH 608
Query 67 IAANHQEDERNFVLSVIENHTQQTEVLDEKQQLLFFEAVGHIISA-TPAEVQASCIEGLM 125
+ + FV ++EN T L Q +F+EAVGHIISA +Q I LM
Sbjct 609 FVIVQPAENKPFVEEMLENLTGIICDLSHAQVHVFYEAVGHIISAQIDGNLQEDLIMKLM 668
Query 126 ANSWEAWQQIVKSGKENQLFLFNPQAAKRLVQILRINQRVAKSTGTAFTPQLSRLYPEMM 185
W I+ + N L P+ K ++ IL+ N KS G++F QL +Y +++
Sbjct 669 DIPNRTWNDIIAAASTNDSVLEEPEMVKSVLNILKTNVAACKSIGSSFVTQLGNIYSDLL 728
Query 186 EIYGVYGQRILQEVAAGGPGRVRHSEIKNLIIFKRETL 223
+Y + +++ + V G +++ +K + KRE L
Sbjct 729 SLYKILSEKVSRAVTTAGEEALKNPLVKTMRAVKREIL 766
> Hs4507943
Length=1071
Score = 167 bits (423), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 88/218 (40%), Positives = 126/218 (57%), Gaps = 1/218 (0%)
Query 7 VVASNIMYVVGQYPRFLRAYWKFLKTVISKLFEFMQEPFPGVRDMACETFLKVANKCKTT 66
++ASNIMY+VGQYPRFLRA+WKFLKTV++KLFEFM E GV+DMAC+TF+K+A KC+
Sbjct 539 IIASNIMYIVGQYPRFLRAHWKFLKTVVNKLFEFMHETHDGVQDMACDTFIKIAQKCRRH 598
Query 67 IAANHQEDERNFVLSVIENHTQQTEVLDEKQQLLFFEAVGHIISA-TPAEVQASCIEGLM 125
+ F+ ++ N L +Q F+EAVG++I A T VQ IE M
Sbjct 599 FVQVQVGEVMPFIDEILNNINTIICDLQPQQVHTFYEAVGYMIGAQTDQTVQEHLIEKYM 658
Query 126 ANSWEAWQQIVKSGKENQLFLFNPQAAKRLVQILRINQRVAKSTGTAFTPQLSRLYPEMM 185
+ W I++ +N L +P+ K+L IL+ N R K+ G F QL R+Y +M+
Sbjct 659 LLPNQVWDSIIQQATKNVDILKDPETVKQLGSILKTNVRACKAVGHPFVIQLGRIYLDML 718
Query 186 EIYGVYGQRILQEVAAGGPGRVRHSEIKNLIIFKRETL 223
+Y + I + A G + I+++ KRETL
Sbjct 719 NVYKCLSENISAAIQANGEMVTKQPLIRSMRTVKRETL 756
> At3g03110
Length=1022
Score = 166 bits (420), Expect = 4e-41, Method: Compositional matrix adjust.
Identities = 89/218 (40%), Positives = 127/218 (58%), Gaps = 1/218 (0%)
Query 7 VVASNIMYVVGQYPRFLRAYWKFLKTVISKLFEFMQEPFPGVRDMACETFLKVANKCKTT 66
V+ASNIMYVVGQY RFLRA+WKFLKTV+ KLFEFM E PGV+DMAC+TFLK+ KCK
Sbjct 489 VIASNIMYVVGQYSRFLRAHWKFLKTVVHKLFEFMHETHPGVQDMACDTFLKIVQKCKRK 548
Query 67 IAANHQEDERNFVLSVIENHTQQTEVLDEKQQLLFFEAVGHIISA-TPAEVQASCIEGLM 125
+ FV ++ L Q F+E+VG +I A + + + ++ LM
Sbjct 549 FVIVQVGESEPFVSELLSGLATIVGDLQPHQIHTFYESVGSMIQAESDPQKRGEYLQRLM 608
Query 126 ANSWEAWQQIVKSGKENQLFLFNPQAAKRLVQILRINQRVAKSTGTAFTPQLSRLYPEMM 185
A + W +I+ +++ L P + ++ IL+ N RVA S GT F Q+S ++ +M+
Sbjct 609 ALPNQKWAEIIGQARQSADILKEPDVIRTVLNILQTNTRVATSLGTFFLSQISLIFLDML 668
Query 186 EIYGVYGQRILQEVAAGGPGRVRHSEIKNLIIFKRETL 223
+Y +Y + + +A GGP R S +K L KRE L
Sbjct 669 NVYRMYSELVSSSIANGGPYASRTSLVKLLRSVKREIL 706
> At5g17020
Length=1075
Score = 164 bits (415), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 87/218 (39%), Positives = 130/218 (59%), Gaps = 1/218 (0%)
Query 7 VVASNIMYVVGQYPRFLRAYWKFLKTVISKLFEFMQEPFPGVRDMACETFLKVANKCKTT 66
V+ASNIMYVVGQYPRFLRA+WKFLKTV++KLFEFM E PGV+DMAC+TFLK+ KCK
Sbjct 542 VIASNIMYVVGQYPRFLRAHWKFLKTVVNKLFEFMHETHPGVQDMACDTFLKIVQKCKRK 601
Query 67 IAANHQEDERNFVLSVIENHTQQTEVLDEKQQLLFFEAVGHIISA-TPAEVQASCIEGLM 125
+ FV ++ + L+ Q F+E+VG++I A + + + ++ LM
Sbjct 602 FVIVQVGENEPFVSELLTGLATTVQDLEPHQIHSFYESVGNMIQAESDPQKRDEYLQRLM 661
Query 126 ANSWEAWQQIVKSGKENQLFLFNPQAAKRLVQILRINQRVAKSTGTAFTPQLSRLYPEMM 185
A + W +I+ + + FL + + ++ IL+ N A S GT F Q+S ++ +M+
Sbjct 662 ALPNQKWAEIIGQARHSVEFLKDQVVIRTVLNILQTNTSAATSLGTYFLSQISLIFLDML 721
Query 186 EIYGVYGQRILQEVAAGGPGRVRHSEIKNLIIFKRETL 223
+Y +Y + + + GGP + S +K L KRETL
Sbjct 722 NVYRMYSELVSTNITEGGPYASKTSFVKLLRSVKRETL 759
> YGR218w
Length=1084
Score = 153 bits (386), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 83/218 (38%), Positives = 120/218 (55%), Gaps = 1/218 (0%)
Query 7 VVASNIMYVVGQYPRFLRAYWKFLKTVISKLFEFMQEPFPGVRDMACETFLKVANKCKTT 66
VVAS+IMYVVGQYPRFL+A+W FL+TVI KLFEFM E GV+DMAC+TF+K+ KCK
Sbjct 550 VVASDIMYVVGQYPRFLKAHWNFLRTVILKLFEFMHETHEGVQDMACDTFIKIVQKCKYH 609
Query 67 IAANHQEDERNFVLSVIENHTQQTEVLDEKQQLLFFEAVGHIISATPAEVQAS-CIEGLM 125
+ F+ ++I + + T L +Q F++A G IIS + + + + LM
Sbjct 610 FVIQQPRESEPFIQTIIRDIQKTTADLQPQQVHTFYKACGIIISEERSVAERNRLLSDLM 669
Query 126 ANSWEAWQQIVKSGKENQLFLFNPQAAKRLVQILRINQRVAKSTGTAFTPQLSRLYPEMM 185
AW IV+ N L + + K + I++ N V S G F PQL +Y M+
Sbjct 670 QLPNMAWDTIVEQSTANPTLLLDSETVKIIANIIKTNVAVCTSMGADFYPQLGHIYYNML 729
Query 186 EIYGVYGQRILQEVAAGGPGRVRHSEIKNLIIFKRETL 223
++Y I +VAA G + +++ L K+E L
Sbjct 730 QLYRAVSSMISAQVAAEGLIATKTPKVRGLRTIKKEIL 767
> ECU11g0240
Length=1058
Score = 89.7 bits (221), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 44/107 (41%), Positives = 65/107 (60%), Gaps = 1/107 (0%)
Query 7 VVASNIMYVVGQYPRFLRAYWKFLKTVISKLFEFMQEPFPGVRDMACETFLKVANKCKTT 66
V+ASNIM+++GQY RFL FLKTV+ KLFEFM E G++DMAC+ F K+ KC
Sbjct 563 VIASNIMFIIGQYHRFLLHNKSFLKTVVKKLFEFMDEDHEGIKDMACDNFFKIVEKCPVE 622
Query 67 IAANHQEDERNFVLSVIENHTQQTEVLDEKQQLLFFEAVGHIISATP 113
+ DE F++ +++N T L+ Q+ + +EA+ +I P
Sbjct 623 FLTQREGDEI-FIVYILKNLPAITGTLEFYQKRIVYEALLMVIREVP 668
> At1g14350
Length=448
Score = 32.7 bits (73), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 22/42 (52%), Gaps = 0/42 (0%)
Query 106 GHIISATPAEVQASCIEGLMANSWEAWQQIVKSGKENQLFLF 147
++S+ +V A E M N+W+ Q + GKEN LF +
Sbjct 239 AELLSSLAQKVNADNTEQSMENAWKVLQDFLNKGKENDLFRY 280
> CE21377
Length=270
Score = 32.7 bits (73), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 37/163 (22%), Positives = 61/163 (37%), Gaps = 25/163 (15%)
Query 47 GVRDMACETFLKVANKCKTTIAANHQEDERNFVLSVIENHTQQTEVLDEKQQLLFFEAVG 106
G R CE + + + + + E +F + Q +D Q ++ V
Sbjct 104 GKRRQICEDKMLMISHGSEYTVMDLTKIEFDFSWCTKYTNEQMQHFIDNTQNSFLYKLVD 163
Query 107 HIISATPAEVQAS------CIE-------GLMANSWEAWQQIVKSGKENQLFLFN----P 149
+++ P++V+ S C G M E +QQ K E F + P
Sbjct 164 YMMDLKPSDVELSFMICQACFHYAGQRFHGKMLEICENFQQ--KLADELHEFYVDEWRMP 221
Query 150 QAAKRLVQILRINQRVAKSTGTAFTPQLSRLYPEMMEIYGVYG 192
+ RL Q+LRIN R+ + Q E+ EI+ VY
Sbjct 222 NYSGRLSQLLRINNRIREDIWKCRKKQ------EIAEIFDVYS 258
> Hs20554806
Length=737
Score = 31.2 bits (69), Expect = 1.7, Method: Composition-based stats.
Identities = 27/103 (26%), Positives = 47/103 (45%), Gaps = 4/103 (3%)
Query 50 DMACETFLKVANKCKTTIAANHQEDERNF-VLSVIENHTQQTEVLDEKQQLLFFEAVGHI 108
+++ T + A K +TIA N E ER F ++++I + + +D L+ +G
Sbjct 633 NVSIPTTIYKAKKIVSTIAINSAEAERGFNLMNIICTRVRNSLTIDHVSDLMTINLLGKE 692
Query 109 IS---ATPAEVQASCIEGLMANSWEAWQQIVKSGKENQLFLFN 148
++ ATP S +A Q+ K ENQL ++N
Sbjct 693 LADWDATPFVKSWSNCNHRLATDTRVRQKSTKVFHENQLAIWN 735
> 7295355
Length=884
Score = 30.8 bits (68), Expect = 2.4, Method: Composition-based stats.
Identities = 33/183 (18%), Positives = 81/183 (44%), Gaps = 22/183 (12%)
Query 7 VVASNIMYVVGQYPRFL--RAYWKFLKTVISKLFEFMQEPFPGVRDMACETFLKVANKCK 64
+V S + ++P+++ + + K+L+ +I +L + + + V++ AC F+ + +
Sbjct 446 LVRSITCWTFMRFPKWVLNQPHDKYLEPLIEELLKCILDSNKRVQEAACSAFVALEEEAC 505
Query 65 TTIAANHQEDERNFVLSVIENHTQQTEVLDEKQQLLFFEAVGHIISATPAEVQASCIEGL 124
T + + + FVL+ + H + ++ + LL E+VGH ++ + I+ L
Sbjct 506 TQLVPYLENMLKTFVLAFSKYHQRNLLIMYDVVGLL-AESVGHHLN------KPQYIDIL 558
Query 125 MANSWEAWQQIVKSGKENQLFLFNPQAAKRLVQILRINQRVAKSTGTAFTPQLSRLYPEM 184
M + W + K+ ++ +L +A + ++F P +Y +
Sbjct 559 MPPLMDKWNLVKDDDKD-------------IIYLLECLSSIATALQSSFLPYCDSVYRKG 605
Query 185 MEI 187
+ I
Sbjct 606 ISI 608
> Hs17433387
Length=237
Score = 30.8 bits (68), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 26/104 (25%), Positives = 44/104 (42%), Gaps = 10/104 (9%)
Query 20 PRF-LRAYWKFLKTVISKLFEFMQEPFPGVRDMACETFLKVANKCKTTIAANHQEDERNF 78
P F L A +L +I M + P + K ++KC+ +A N ED + +
Sbjct 37 PGFELPAAQDWLAQLIQSYLYDMSKAHPH------DDVFKYSHKCEDKLALNKTEDSKEY 90
Query 79 VLSVIENHTQQTEVL--DEKQQLLFFEAVGHIISATPAEVQASC 120
+ V+ H Q + + D++ ++F E G II A C
Sbjct 91 NIGVLR-HLQNLDPIAADQQDTVVFKEEEGKIIRALSDPRAGHC 133
> Hs17433389
Length=285
Score = 30.4 bits (67), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 26/104 (25%), Positives = 44/104 (42%), Gaps = 10/104 (9%)
Query 20 PRF-LRAYWKFLKTVISKLFEFMQEPFPGVRDMACETFLKVANKCKTTIAANHQEDERNF 78
P F L A +L +I M + P + K ++KC+ +A N ED + +
Sbjct 85 PGFELPAAQDWLAQLIQSYLYDMSKAHPH------DDVFKYSHKCEDKLALNKTEDSKEY 138
Query 79 VLSVIENHTQQTEVL--DEKQQLLFFEAVGHIISATPAEVQASC 120
+ V+ H Q + + D++ ++F E G II A C
Sbjct 139 NIGVLR-HLQNLDPIAADQQDTVVFKEEEGKIIRALSDPRAGHC 181
> At2g47030
Length=588
Score = 30.0 bits (66), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 20/70 (28%), Positives = 34/70 (48%), Gaps = 5/70 (7%)
Query 102 FEAVGHIISATPAEVQASCIEGLMANSWEAWQQIVKSGKENQLFLFNPQAAKRLVQILRI 161
F+ V + A P + CI + A + +Q++ K+N +F+F A K ++
Sbjct 287 FKTVQQAVDACPENNRGRCIIYIKAGLYR--EQVIIPKKKNNIFMFGDGARK---TVISY 341
Query 162 NQRVAKSTGT 171
N+ VA S GT
Sbjct 342 NRSVALSRGT 351
> At2g02820
Length=446
Score = 29.6 bits (65), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 19/69 (27%), Positives = 32/69 (46%), Gaps = 7/69 (10%)
Query 108 IISATPAEVQASCIEGLMANSWEAWQQIVKSGKENQLFL-------FNPQAAKRLVQILR 160
++S+ +V A + M N+W+ Q + KEN LF F K LV+ LR
Sbjct 253 LLSSLAQKVNADNTDQSMENAWKVLQDFLNKSKENDLFRYGIPDIDFQLDEFKDLVEDLR 312
Query 161 INQRVAKST 169
+ ++S+
Sbjct 313 SSNEDSQSS 321
> 7295587
Length=2110
Score = 29.6 bits (65), Expect = 5.5, Method: Composition-based stats.
Identities = 13/53 (24%), Positives = 30/53 (56%), Gaps = 0/53 (0%)
Query 112 TPAEVQASCIEGLMANSWEAWQQIVKSGKENQLFLFNPQAAKRLVQILRINQR 164
T E+++SCI G + + +W+Q+ K + L +++ + AK ++ ++ R
Sbjct 903 TEIEIKSSCINGKRMSEYRSWRQVKLEIKGDLLRIYSGRHAKSEHNVIELDIR 955
> CE06100
Length=694
Score = 29.6 bits (65), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 26/123 (21%), Positives = 57/123 (46%), Gaps = 5/123 (4%)
Query 25 AYWKFLKTVISKLFEFMQEPFPGVRDMACETFLKVANKCKTTIAANHQEDERNFVLSVIE 84
++ ++ + LF+ + E P VRD+ CET L + + ++R +++V+
Sbjct 208 SFCNYISEISDGLFKMLGEQAPAVRDL-CETVLGNFLTAIRSKPESLSLEDRIQMINVLV 266
Query 85 NHTQQTE-VLDEKQQLLFFEAVGHIISATPAEVQASCIEGLMANSWEAWQQIVKSGKENQ 143
HT + E L K L++ E + + ++C+ G++ + E +K+ N+
Sbjct 267 VHTHENEPFLARKLSLIWLEEFVKLYKTDLLVMLSTCLVGILPSIVETE---LKADAVNR 323
Query 144 LFL 146
L +
Sbjct 324 LLM 326
Lambda K H
0.322 0.135 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4255059914
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40