bitscore colors: <40, 40-50 , 50-80, 80-200, >200
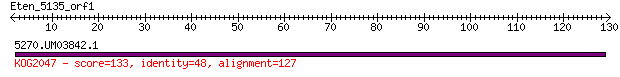
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggV2
2,483,276 sequences; 915,453,621 total letters
Query= Eten_5135_orf1
Length=129
Score E
Sequences producing significant alignments: (Bits) Value
5270.UM03842.1 133 1e-30
> 5270.UM03842.1
Length=1081
Score = 133 bits (335), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 61/133 (45%), Positives = 91/133 (68%), Gaps = 6/133 (4%)
Query 2 DLENAAKVFERAVQATYRTIDDLASVWCEAVEMHLRHGLFKEALRLVRLAISSSR--KG- 58
DLE+A K+FE+A+ +R +DDLA +WCE EM LRH + EA+R + +++ R KG
Sbjct 657 DLESARKIFEKAITIPFRRVDDLAEIWCEWAEMELRHSNYDEAIRTMARSVAPPRNTKGI 716
Query 59 ---QDTKGAQGRLFRSVKLWSLAADVEEMFGSIETVRACYDKMFQLKVITPQLVINYAHY 115
DT Q RLF+S+KLWS D+EE G +E+ + Y+KM +LK+ + Q++INYA +
Sbjct 717 QYHDDTLPPQTRLFKSLKLWSFYVDLEESLGDVESTKRVYEKMLELKIASAQIIINYAAF 776
Query 116 LEERRYYEESFRV 128
LE+ +Y+EESF+V
Sbjct 777 LEDNKYFEESFKV 789
Lambda K H
0.324 0.135 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 22342951125
Database: eggV2
Posted date: Dec 15, 2009 4:47 PM
Number of letters in database: 915,453,621
Number of sequences in database: 2,483,276
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40