bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggV2
2,483,276 sequences; 915,453,621 total letters
Query= Eten_1910_orf3
Length=552
Score E
Sequences producing significant alignments: (Bits) Value
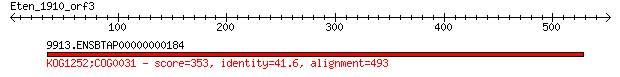
9913.ENSBTAP00000000184 353 9e-96
> 9913.ENSBTAP00000000184
Length=492
Score = 353 bits (906), Expect = 9e-96, Method: Compositional matrix adjust.
Identities = 205/506 (40%), Positives = 288/506 (56%), Gaps = 64/506 (12%)
Query 35 LPSILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRL 94
LP IL IG TP+V ++++G G+ C+LL KCEF +AGGS+KDRI+ MI AE G +
Sbjct 7 LPDILKKIGDTPMVRINKIGRNFGLKCELLAKCEFFNAGGSVKDRISLRMIEDAEREGTI 66
Query 95 APGAALIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTPTKAA 154
PG +IEPTSGNTGIG+AL AAVKGYR + VMP+KMS EK ++RALG+ I+RTPT A
Sbjct 67 KPGDTIIEPTSGNTGIGLALAAAVKGYRCIIVMPEKMSTEKVDVLRALGAEIVRTPTNAR 126
Query 155 WDDQTSHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANP 214
+D SH+ ++ RL+ E+ ILDQYRN +NP
Sbjct 127 FDSPESHVGVAWRLRNEIPNSH---------------------------ILDQYRNASNP 159
Query 215 ISHFFGTGPELLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDPEGSILAN 274
++H+ T E+L QC +LDM+V GTGG++TG+ R++K P C +V VDPEGSILA
Sbjct 160 LAHYDITAEEILQQCDGRLDMLVASAGTGGTITGVARKLKEKCPGCKIVGVDPEGSILAE 219
Query 275 P---NEQPGKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLIRLEGMLVGGS 331
P N+ Y VEGIGYDF+PTVLDR V D W K ND E+ +R+LI EG+L GGS
Sbjct 220 PEELNQTEQTAYEVEGIGYDFIPTVLDRTVVDKWFKSNDEEAFAFARMLIAQEGLLCGGS 279
Query 332 SGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRNYTSKFVCDEWMAEKGFLERSELSRQ 391
SG+A++ A+KA + + +R ++LPD RNY SKF+ D+WM +KGF++ E+S +
Sbjct 280 SGSAMSVAVKAAQEL----QEGQRCVVILPDSVRNYMSKFLSDKWMLQKGFMKEEEISVK 335
Query 392 YEQYSNMSVQSLQ-------LPRLTFVSSDSSLKEAAALLVQSPQPILAVIEPGAAEDAT 444
+ ++ VQ L LP +T + L+E Q+P V E G
Sbjct 336 RPWWWHLQVQELSLSAPLTVLPTVTCEHTIEILREKG--FDQAP----VVDESG------ 383
Query 445 QNGTAPRQMVTGSVTQKALLMAL--GEMPSSAPIGEIADTETPVYNIGTPLSLLAQSLEL 502
++ G VT +L +L G++ S + ++ + + PL L+ LE+
Sbjct 384 --------VILGMVTLGNMLSSLLAGKVQPSDQVCKVIYKQFKQIYLTDPLGKLSHILEM 435
Query 503 HPFLFIESDGI-IRAAVESSQLLQAF 527
F + + I R ESS+ F
Sbjct 436 DHFALVVHEQIQYRCHGESSKRQMVF 461
Lambda K H
0.316 0.132 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 245189545547
Database: eggV2
Posted date: Dec 15, 2009 4:47 PM
Number of letters in database: 915,453,621
Number of sequences in database: 2,483,276
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40