bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggV2
2,483,276 sequences; 915,453,621 total letters
Query= Eten_0308_orf10
Length=697
Score E
Sequences producing significant alignments: (Bits) Value
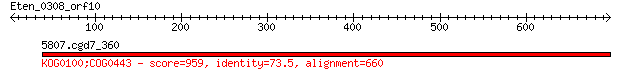
5807.cgd7_360 959 0.0
> 5807.cgd7_360
Length=655
Score = 959 bits (2480), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 485/660 (73%), Positives = 574/660 (86%), Gaps = 10/660 (1%)
Query 38 ALPFCFFLLSLFSHYPHAVRGDDTEGDGKVKDVVIGIDLGTTYSCVGVYRQGRVDIIPND 97
A+ F FL SLF +P VR D + K VIGIDLGTTYSCVG+Y+ GRV+IIPN+
Sbjct 6 AIIFSVFL-SLFLAFP--VRSSD---EKKYDGPVIGIDLGTTYSCVGIYKNGRVEIIPNE 59
Query 98 QGNRITPSYVSFAEDERKIGEAAKNEAAINPTNTIFDVKRLIGRRFNEKEVQRDKELLPY 157
QGNRITPSYVSF +DER IGE+AKN+A INP T+FDVKRLIGRRF + VQ+DK LLPY
Sbjct 60 QGNRITPSYVSFTDDERLIGESAKNQATINPVQTLFDVKRLIGRRFKDDSVQKDKTLLPY 119
Query 158 EIINKDGKPYIRVMVKGQPKELAPEEVSAMVLGKMKEVAESYLGKEVKNAVVTVPAYFND 217
EIINKD KPYI+V VKG+ K+LAPEEVSAMVL KMKE+AE+YLGKEVK+AV+TVPAYFND
Sbjct 120 EIINKDSKPYIQVSVKGEKKQLAPEEVSAMVLVKMKEIAEAYLGKEVKHAVITVPAYFND 179
Query 218 AQRQATKDAGAIAGLNVIRIINEPTAAAIAYGLDKKDEKTILVYDLGGGTFDVSVLVIDN 277
AQRQATKDAGAIAGLNVIRIINEPTAAAIA+GLDKK EK+ILVYDLGGGTFDVS+L IDN
Sbjct 180 AQRQATKDAGAIAGLNVIRIINEPTAAAIAFGLDKKAEKSILVYDLGGGTFDVSLLTIDN 239
Query 278 GVFEVHATSGDTHLGGEDFDQRVMDHFLKIIQKKYNKDLRKDKSALQRLRREVERAKRAL 337
GVFEV ATSGDTHLGGEDFDQRVMDHF+KII+ K KD++ DK ALQ+LRREVE++KRAL
Sbjct 240 GVFEVVATSGDTHLGGEDFDQRVMDHFIKIIKSKTGKDVKSDKRALQKLRREVEKSKRAL 299
Query 338 SSSHQVTVEVEGLVDGEDFSETLTRAKFEELNADLFQKTLKPVKQVLEDADLKKSQIDEI 397
SS+ QV VE+EGL++ D SETLTRAKF+ELNADLF+KT++PVK+VLEDA +KKS++DEI
Sbjct 300 SSAPQVKVEIEGLMEDVDLSETLTRAKFDELNADLFRKTIEPVKKVLEDAGIKKSEVDEI 359
Query 398 VLVGGSTRIPKIQQLIKEFFNGKEPNRGINPDEAVAYGAAVQAGILSGEGTQDMVLLDVT 457
VLVGGSTRIPKIQ LIKEFF+GKEPNRGINPDEAVAYGAAVQAGIL+GEG D++LLDVT
Sbjct 360 VLVGGSTRIPKIQALIKEFFDGKEPNRGINPDEAVAYGAAVQAGILAGEGGSDLLLLDVT 419
Query 458 PLTLGIETAGGVMAKVINKNTVIPTKKTQTFSTYSDNQSAVLIQVYEGERPMTKNNHLLG 517
PLTLGIET GGVM K+I +NTV+PTKK+Q FSTY DNQ AVLIQVYEGERPMTK+N+LLG
Sbjct 420 PLTLGIETVGGVMTKLIGRNTVVPTKKSQVFSTYQDNQPAVLIQVYEGERPMTKDNNLLG 479
Query 518 KFELTGIPPAPRGVPQIDVTFDVDRNGILNVSAVDKGTGKSEKITITNDKGRLTPDEIER 577
KFEL+GIPPAPRGVPQI+VTF++D +GIL VSA DKGTGKSEKITITNDKGRL+ ++IER
Sbjct 480 KFELSGIPPAPRGVPQIEVTFEIDTDGILQVSAKDKGTGKSEKITITNDKGRLSQEDIER 539
Query 578 MIQEAERFADEDRKTKERVDARNALEGYLHSMRSTVEDKDKLADKIEDDDKKTIMDKITE 637
MI+EAE+FA+ED+ +E+VDA+NAL+ Y+HSMR ++EDKDKLA K+E++DK+ I + + +
Sbjct 540 MIKEAEQFAEEDKLVREKVDAKNALDSYVHSMRMSIEDKDKLAQKLEEEDKEKIKEALKD 599
Query 638 ANEWLVANPEADGEELRDKLKDVESVCNPIISKVYGQTGGPSDSGATSTSDDDYSSHDEL 697
A ++L +NP+AD +E++DKLK+VE +CNPII+ VYGQ + A DDYS HDEL
Sbjct 600 AEDFLSSNPDADAQEIKDKLKEVEGICNPIIAAVYGQ----AGGAAGHAGGDDYSGHDEL 655
Lambda K H
0.314 0.134 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 326078784882
Database: eggV2
Posted date: Dec 15, 2009 4:47 PM
Number of letters in database: 915,453,621
Number of sequences in database: 2,483,276
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40