bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_3393_orf1
Length=104
Score E
Sequences producing significant alignments: (Bits) Value
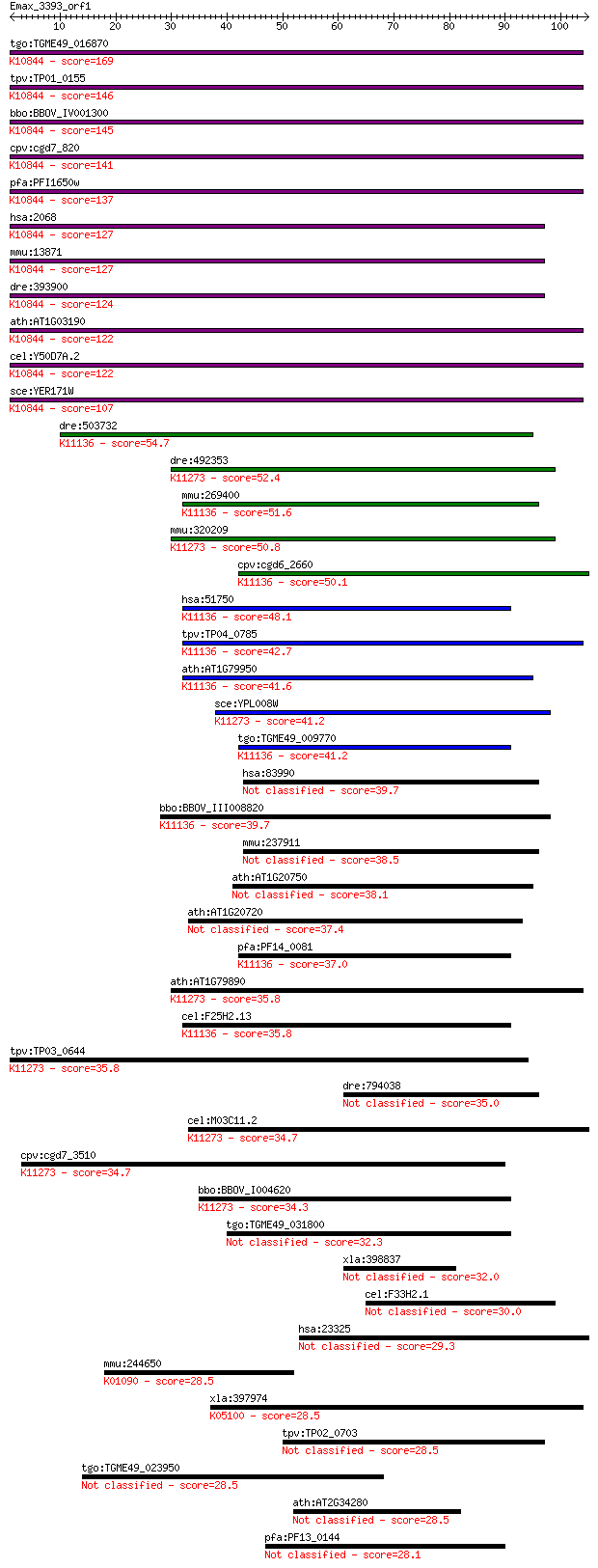
tgo:TGME49_016870 excision repair protein rad15, putative ; K1... 169 3e-42
tpv:TP01_0155 DNA repair protein Rad3; K10844 DNA excision rep... 146 2e-35
bbo:BBOV_IV001300 21.m02831; DNA excision repair helicase; K10... 145 4e-35
cpv:cgd7_820 RAD3'DEXDc+HELICc protein' ; K10844 DNA excision ... 141 6e-34
pfa:PFI1650w DNA excision-repair helicase, putative; K10844 DN... 137 6e-33
hsa:2068 ERCC2, COFS2, EM9, MGC102762, MGC126218, MGC126219, T... 127 8e-30
mmu:13871 Ercc2, AA407812, AU020867, AW240756, Ercc-2, XPD; ex... 127 1e-29
dre:393900 ercc2, MGC56365, zgc:56365; excision repair cross-c... 124 5e-29
ath:AT1G03190 UVH6; UVH6 (ULTRAVIOLET HYPERSENSITIVE 6); ATP b... 122 3e-28
cel:Y50D7A.2 hypothetical protein; K10844 DNA excision repair ... 122 3e-28
sce:YER171W RAD3, REM1; 5' to 3' DNA helicase, involved in nuc... 107 6e-24
dre:503732 rtel1; zgc:113114 (EC:3.6.4.12); K11136 regulator o... 54.7 8e-08
dre:492353 zgc:92172 (EC:3.6.4.13); K11273 chromosome transmis... 52.4 4e-07
mmu:269400 Rtel1, AI451565, AW540478, Rtel; regulator of telom... 51.6 6e-07
mmu:320209 Ddx11, 4732462I11Rik, CHL1, CHLR1, KRG2, MGC90809; ... 50.8 9e-07
cpv:cgd6_2660 DNA repair helicase ; K11136 regulator of telome... 50.1 2e-06
hsa:51750 RTEL1, C20orf41, DKFZp434C013, KIAA1088, NHL, RTEL, ... 48.1 6e-06
tpv:TP04_0785 DNA repair helicase; K11136 regulator of telomer... 42.7 2e-04
ath:AT1G79950 helicase-related; K11136 regulator of telomere e... 41.6 6e-04
sce:YPL008W CHL1, CTF1, LPA9, MCM12; Chl1p (EC:3.6.1.-); K1127... 41.2 9e-04
tgo:TGME49_009770 DNA repair helicase, putative ; K11136 regul... 41.2 0.001
hsa:83990 BRIP1, BACH1, FANCJ, FLJ90232, MGC126521, MGC126523,... 39.7 0.003
bbo:BBOV_III008820 17.m07769; DNA repair helicase (rad3) famil... 39.7 0.003
mmu:237911 Brip1, 3110009N10Rik, 8030460J03Rik, Bach1, FACJ, F... 38.5 0.006
ath:AT1G20750 helicase-related 38.1 0.006
ath:AT1G20720 ATP binding / ATP-dependent DNA helicase/ ATP-de... 37.4 0.012
pfa:PF14_0081 DNA-repair helicase, putative; K11136 regulator ... 37.0 0.016
ath:AT1G79890 helicase-related; K11273 chromosome transmission... 35.8 0.030
cel:F25H2.13 rtel-1; RTEL (mammalian Regulator of TELomere len... 35.8 0.032
tpv:TP03_0644 DNA helicase; K11273 chromosome transmission fid... 35.8 0.038
dre:794038 brip1, fancj, si:ch211-158l18.1; BRCA1 interacting ... 35.0 0.052
cel:M03C11.2 hypothetical protein; K11273 chromosome transmiss... 34.7 0.066
cpv:cgd7_3510 helicase ; K11273 chromosome transmission fideli... 34.7 0.076
bbo:BBOV_I004620 19.m02044; DNA repair helicase (rad3) and DEA... 34.3 0.091
tgo:TGME49_031800 hypothetical protein 32.3 0.40
xla:398837 brip1, MGC68622; BRCA1 interacting protein C-termin... 32.0 0.45
cel:F33H2.1 dog-1; Deletions Of G-rich DNA family member (dog-1) 30.0 2.1
hsa:23325 KIAA1033 29.3 3.2
mmu:244650 Phlpp2, AI481772, C130044A18Rik, KIAA0931, Phlppl; ... 28.5 4.7
xla:397974 mst1r, Xhlp, Xron; macrophage stimulating 1 recepto... 28.5 5.3
tpv:TP02_0703 hypothetical protein 28.5 5.5
tgo:TGME49_023950 hypothetical protein 28.5 5.6
ath:AT2G34280 S locus F-box-related / SLF-related 28.5 6.0
pfa:PF13_0144 oxidoreductase, putative (EC:1.1.1.-) 28.1 7.1
> tgo:TGME49_016870 excision repair protein rad15, putative ;
K10844 DNA excision repair protein ERCC-2 [EC:3.6.4.12]
Length=1065
Score = 169 bits (427), Expect = 3e-42, Method: Compositional matrix adjust.
Identities = 73/103 (70%), Positives = 91/103 (88%), Gaps = 0/103 (0%)
Query 1 PLELYPKILSFVPLIAESFPVSMERACFCPMIVARGADQVPLTSKFECRHDVSVLRNYAN 60
PLELYPK+L+FVP+I ESFP+S++R C CP+IVARG+DQ+PLTSKFE RHD++VLRNY+N
Sbjct 733 PLELYPKLLNFVPVITESFPMSLDRNCICPLIVARGSDQIPLTSKFEYRHDMNVLRNYSN 792
Query 61 LLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGFLAQTLPFK 103
LL+DLCK VPDGLVCFF SY+YME+ +S WY +G LAQ L +K
Sbjct 793 LLIDLCKHVPDGLVCFFTSYSYMESVLSSWYHSGVLAQVLDYK 835
> tpv:TP01_0155 DNA repair protein Rad3; K10844 DNA excision repair
protein ERCC-2 [EC:3.6.4.12]
Length=894
Score = 146 bits (368), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 59/103 (57%), Positives = 84/103 (81%), Gaps = 0/103 (0%)
Query 1 PLELYPKILSFVPLIAESFPVSMERACFCPMIVARGADQVPLTSKFECRHDVSVLRNYAN 60
PLE YPKIL+F P++ +S P+S++R C CP+IV++GA+QV +++KF+ R D++VLRNY +
Sbjct 599 PLEFYPKILNFTPVLMQSLPMSLDRDCLCPIIVSKGANQVHMSTKFDLRTDITVLRNYGS 658
Query 61 LLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGFLAQTLPFK 103
L++DLCK +PDG+VCFFPSYAYME +S+WYE G LA + K
Sbjct 659 LVIDLCKHIPDGVVCFFPSYAYMELILSHWYETGILASIMEHK 701
> bbo:BBOV_IV001300 21.m02831; DNA excision repair helicase; K10844
DNA excision repair protein ERCC-2 [EC:3.6.4.12]
Length=822
Score = 145 bits (365), Expect = 4e-35, Method: Compositional matrix adjust.
Identities = 58/103 (56%), Positives = 85/103 (82%), Gaps = 0/103 (0%)
Query 1 PLELYPKILSFVPLIAESFPVSMERACFCPMIVARGADQVPLTSKFECRHDVSVLRNYAN 60
PLE+YPKIL+F P++ +S P+S++R C CP+IVA+GA+Q+ +++++E R+DV+VLRNY
Sbjct 525 PLEMYPKILNFTPVLTQSLPMSLDRDCLCPLIVAKGANQLQMSTRYELRNDVTVLRNYGT 584
Query 61 LLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGFLAQTLPFK 103
LL++LCK +PDG+VCFFPSYAYME +S+WYE G +A + K
Sbjct 585 LLIELCKHIPDGVVCFFPSYAYMELIVSHWYECGIIASIMEHK 627
> cpv:cgd7_820 RAD3'DEXDc+HELICc protein' ; K10844 DNA excision
repair protein ERCC-2 [EC:3.6.4.12]
Length=841
Score = 141 bits (355), Expect = 6e-34, Method: Composition-based stats.
Identities = 57/103 (55%), Positives = 80/103 (77%), Gaps = 0/103 (0%)
Query 1 PLELYPKILSFVPLIAESFPVSMERACFCPMIVARGADQVPLTSKFECRHDVSVLRNYAN 60
PL+LYPK+L F+P++++S ++++R C CP+IV RG+DQ PL+SKFE R DVS+ +NY
Sbjct 565 PLDLYPKLLGFIPVVSQSLTMTLDRTCICPLIVTRGSDQTPLSSKFESRADVSIQQNYGK 624
Query 61 LLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGFLAQTLPFK 103
L+L++ K VPDG+VCFF SY YME +S WYE+G LAQ + K
Sbjct 625 LILEITKKVPDGVVCFFSSYLYMEQMLSQWYESGLLAQIMEHK 667
> pfa:PFI1650w DNA excision-repair helicase, putative; K10844
DNA excision repair protein ERCC-2 [EC:3.6.4.12]
Length=1056
Score = 137 bits (346), Expect = 6e-33, Method: Composition-based stats.
Identities = 55/103 (53%), Positives = 79/103 (76%), Gaps = 0/103 (0%)
Query 1 PLELYPKILSFVPLIAESFPVSMERACFCPMIVARGADQVPLTSKFECRHDVSVLRNYAN 60
PLELYPK+L+F ++ SFP+S +R C CP+IV +G+D +PL+S+F R+D+SV++NY
Sbjct 758 PLELYPKLLNFKTVLTASFPMSFDRNCVCPLIVTKGSDLIPLSSQFSLRNDLSVIKNYGI 817
Query 61 LLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGFLAQTLPFK 103
LL+D+CK +PDG+V +FPSY YME +S WYE G +A L +K
Sbjct 818 LLVDMCKCIPDGIVAYFPSYIYMEQVISSWYELGVIANILEYK 860
> hsa:2068 ERCC2, COFS2, EM9, MGC102762, MGC126218, MGC126219,
TTD, XPD; excision repair cross-complementing rodent repair
deficiency, complementation group 2 (EC:3.6.4.12); K10844 DNA
excision repair protein ERCC-2 [EC:3.6.4.12]
Length=760
Score = 127 bits (319), Expect = 8e-30, Method: Composition-based stats.
Identities = 52/96 (54%), Positives = 71/96 (73%), Gaps = 0/96 (0%)
Query 1 PLELYPKILSFVPLIAESFPVSMERACFCPMIVARGADQVPLTSKFECRHDVSVLRNYAN 60
PL++YPKIL F P+ +F +++ R C CPMI+ RG DQV ++SKFE R D++V+RNY N
Sbjct 463 PLDIYPKILDFHPVTMATFTMTLARVCLCPMIIGRGNDQVAISSKFETREDIAVIRNYGN 522
Query 61 LLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGFL 96
LLL++ VPDG+V FF SY YME+ ++ WYE G L
Sbjct 523 LLLEMSAVVPDGIVAFFTSYQYMESTVASWYEQGIL 558
> mmu:13871 Ercc2, AA407812, AU020867, AW240756, Ercc-2, XPD;
excision repair cross-complementing rodent repair deficiency,
complementation group 2 (EC:3.6.4.12); K10844 DNA excision
repair protein ERCC-2 [EC:3.6.4.12]
Length=760
Score = 127 bits (318), Expect = 1e-29, Method: Composition-based stats.
Identities = 52/96 (54%), Positives = 71/96 (73%), Gaps = 0/96 (0%)
Query 1 PLELYPKILSFVPLIAESFPVSMERACFCPMIVARGADQVPLTSKFECRHDVSVLRNYAN 60
PL++YPKIL F P+ +F +++ R C CPMI+ RG DQV ++SKFE R D++V+RNY N
Sbjct 463 PLDIYPKILDFHPVTMATFTMTLARVCLCPMIIGRGNDQVAISSKFETREDIAVIRNYGN 522
Query 61 LLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGFL 96
LLL++ VPDG+V FF SY YME+ ++ WYE G L
Sbjct 523 LLLEMSAVVPDGIVAFFTSYQYMESTVASWYEQGIL 558
> dre:393900 ercc2, MGC56365, zgc:56365; excision repair cross-complementing
rodent repair deficiency, complementation group
2; K10844 DNA excision repair protein ERCC-2 [EC:3.6.4.12]
Length=643
Score = 124 bits (312), Expect = 5e-29, Method: Compositional matrix adjust.
Identities = 53/96 (55%), Positives = 69/96 (71%), Gaps = 0/96 (0%)
Query 1 PLELYPKILSFVPLIAESFPVSMERACFCPMIVARGADQVPLTSKFECRHDVSVLRNYAN 60
PL++YP+IL F P+ SF +++ R C CP+IV RG DQV +TSKFE R D +V+RNY N
Sbjct 463 PLDIYPRILDFRPVTMASFTMTLARTCLCPLIVGRGNDQVAMTSKFETREDFAVIRNYGN 522
Query 61 LLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGFL 96
LLL++ VPDG+V FF SY YME ++ WYE G L
Sbjct 523 LLLEMSAIVPDGIVAFFTSYMYMENIVASWYEQGIL 558
> ath:AT1G03190 UVH6; UVH6 (ULTRAVIOLET HYPERSENSITIVE 6); ATP
binding / ATP-dependent DNA helicase/ ATP-dependent helicase/
DNA binding / hydrolase, acting on acid anhydrides, in phosphorus-containing
anhydrides / nucleic acid binding; K10844
DNA excision repair protein ERCC-2 [EC:3.6.4.12]
Length=758
Score = 122 bits (306), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 48/103 (46%), Positives = 76/103 (73%), Gaps = 0/103 (0%)
Query 1 PLELYPKILSFVPLIAESFPVSMERACFCPMIVARGADQVPLTSKFECRHDVSVLRNYAN 60
P++LYP++L+F P+++ SF +SM R C CPM++ RG+DQ+P+++KF+ R D V+RNY
Sbjct 463 PIDLYPRLLNFTPVVSRSFKMSMTRDCICPMVLTRGSDQLPVSTKFDMRSDPGVVRNYGK 522
Query 61 LLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGFLAQTLPFK 103
LL+++ VPDG+VCFF SY+YM+ ++ W E G L + + K
Sbjct 523 LLVEMVSIVPDGVVCFFVSYSYMDGIIATWNETGILKEIMQQK 565
> cel:Y50D7A.2 hypothetical protein; K10844 DNA excision repair
protein ERCC-2 [EC:3.6.4.12]
Length=606
Score = 122 bits (305), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 51/103 (49%), Positives = 72/103 (69%), Gaps = 0/103 (0%)
Query 1 PLELYPKILSFVPLIAESFPVSMERACFCPMIVARGADQVPLTSKFECRHDVSVLRNYAN 60
PLE+YPK+L F P + SF +++ R C P++VARG DQV +TS+FE R DV+V+RNY N
Sbjct 308 PLEMYPKVLDFDPSVIASFTMTLARPCLSPLVVARGNDQVAMTSRFEQRADVAVIRNYGN 367
Query 61 LLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGFLAQTLPFK 103
L+L++ VPDG+V FF SY YME + WYE + + + +K
Sbjct 368 LVLEMASLVPDGMVVFFTSYLYMENVIGVWYEQHIIDELMKYK 410
> sce:YER171W RAD3, REM1; 5' to 3' DNA helicase, involved in nucleotide
excision repair and transcription; subunit of RNA
polymerase II transcription initiation factor TFIIH; subunit
of Nucleotide Excision Repair Factor 3 (NEF3); homolog of human
XPD protein (EC:3.6.1.-); K10844 DNA excision repair protein
ERCC-2 [EC:3.6.4.12]
Length=778
Score = 107 bits (268), Expect = 6e-24, Method: Composition-based stats.
Identities = 43/103 (41%), Positives = 73/103 (70%), Gaps = 0/103 (0%)
Query 1 PLELYPKILSFVPLIAESFPVSMERACFCPMIVARGADQVPLTSKFECRHDVSVLRNYAN 60
PL++YP++L+F ++ +S+ +++ + F PMI+ +G+DQV ++S+FE R+D S++RNY +
Sbjct 465 PLDMYPRMLNFKTVLQKSYAMTLAKKSFLPMIITKGSDQVAISSRFEIRNDPSIVRNYGS 524
Query 61 LLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGFLAQTLPFK 103
+L++ K PDG+V FFPSY YME+ +S W G L + K
Sbjct 525 MLVEFAKITPDGMVVFFPSYLYMESIVSMWQTMGILDEVWKHK 567
> dre:503732 rtel1; zgc:113114 (EC:3.6.4.12); K11136 regulator
of telomere elongation helicase 1
Length=1177
Score = 54.7 bits (130), Expect = 8e-08, Method: Composition-based stats.
Identities = 30/91 (32%), Positives = 46/91 (50%), Gaps = 6/91 (6%)
Query 10 SFVPLIAESFPVSME------RACFCPMIVARGADQVPLTSKFECRHDVSVLRNYANLLL 63
SF + FPVS+E R I+ +G D V L++ F+ R + + N L+
Sbjct 486 SFTCEMQIPFPVSLENPHVIQRDQIFVSIIEKGPDGVQLSTAFDRRFVPENMSSMGNTLV 545
Query 64 DLCKSVPDGLVCFFPSYAYMETAMSYWYENG 94
+L + VP GL+ FFPSY M+ + +W G
Sbjct 546 NLSRVVPHGLLVFFPSYPVMDKTLEFWRAKG 576
> dre:492353 zgc:92172 (EC:3.6.4.13); K11273 chromosome transmission
fidelity protein 1 [EC:3.6.4.13]
Length=890
Score = 52.4 bits (124), Expect = 4e-07, Method: Composition-based stats.
Identities = 22/69 (31%), Positives = 35/69 (50%), Gaps = 0/69 (0%)
Query 30 PMIVARGADQVPLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSY 89
P+++ G L F+ R ++ +L +LC VP G+VCFFPSY Y + + +
Sbjct 643 PIVLCAGPSGQQLEFTFQTRDSPQMMEETGRVLSNLCNIVPGGVVCFFPSYEYEKRILGH 702
Query 90 WYENGFLAQ 98
W G L +
Sbjct 703 WESTGILQR 711
> mmu:269400 Rtel1, AI451565, AW540478, Rtel; regulator of telomere
elongation helicase 1 (EC:3.6.4.12); K11136 regulator
of telomere elongation helicase 1
Length=1209
Score = 51.6 bits (122), Expect = 6e-07, Method: Composition-based stats.
Identities = 24/64 (37%), Positives = 35/64 (54%), Gaps = 0/64 (0%)
Query 32 IVARGADQVPLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYWY 91
IV RG D V L+S ++ R L + L ++ + VP GL+ FFPSY ME ++ +W
Sbjct 515 IVPRGPDGVQLSSAYDKRFSEECLSSLGKALSNIARVVPHGLLVFFPSYPVMEKSLEFWQ 574
Query 92 ENGF 95
G
Sbjct 575 VQGL 578
> mmu:320209 Ddx11, 4732462I11Rik, CHL1, CHLR1, KRG2, MGC90809;
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 11 (CHL1-like
helicase homolog, S. cerevisiae) (EC:3.6.4.13); K11273 chromosome
transmission fidelity protein 1 [EC:3.6.4.13]
Length=880
Score = 50.8 bits (120), Expect = 9e-07, Method: Composition-based stats.
Identities = 21/69 (30%), Positives = 35/69 (50%), Gaps = 0/69 (0%)
Query 30 PMIVARGADQVPLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSY 89
P+I+ G L ++ R ++ +L +LC VP G+VCF PSY Y+ ++
Sbjct 632 PLIICSGPSNQQLEFTYQRRELPQMVEETGRILCNLCNVVPGGVVCFLPSYEYLRQVHAH 691
Query 90 WYENGFLAQ 98
W + G L +
Sbjct 692 WDKTGLLTR 700
> cpv:cgd6_2660 DNA repair helicase ; K11136 regulator of telomere
elongation helicase 1
Length=1108
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 19/63 (30%), Positives = 37/63 (58%), Gaps = 0/63 (0%)
Query 42 LTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGFLAQTLP 101
L +E R++ S + +++ D K +PDG++ FF SY+ M+ A+ +W + G + +
Sbjct 611 LIGSYEARNNPSYFSSLGSVVFDCVKRIPDGILLFFGSYSLMDQAVKHWTDQGLIERIKA 670
Query 102 FKA 104
FK+
Sbjct 671 FKS 673
> hsa:51750 RTEL1, C20orf41, DKFZp434C013, KIAA1088, NHL, RTEL,
bK3184A7.3; regulator of telomere elongation helicase 1 (EC:3.6.4.12);
K11136 regulator of telomere elongation helicase
1
Length=1219
Score = 48.1 bits (113), Expect = 6e-06, Method: Composition-based stats.
Identities = 22/59 (37%), Positives = 33/59 (55%), Gaps = 0/59 (0%)
Query 32 IVARGADQVPLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYW 90
+V RG D L+S F+ R L + L ++ + VP GL+ FFPSY ME ++ +W
Sbjct 515 VVPRGPDGAQLSSAFDRRFSEECLSSLGKALGNIARVVPYGLLIFFPSYPVMEKSLEFW 573
> tpv:TP04_0785 DNA repair helicase; K11136 regulator of telomere
elongation helicase 1
Length=962
Score = 42.7 bits (99), Expect = 2e-04, Method: Composition-based stats.
Identities = 22/72 (30%), Positives = 35/72 (48%), Gaps = 0/72 (0%)
Query 32 IVARGADQVPLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYWY 91
I D L+S F R+ ++ + N +L K+VP G++ FF SY+ M S W
Sbjct 599 ISGNAEDPNMLSSTFNTRNKMNYITELGNAVLSFVKNVPGGVLVFFGSYSVMNYTASVWK 658
Query 92 ENGFLAQTLPFK 103
+ G ++ FK
Sbjct 659 KIGIYSKIEMFK 670
> ath:AT1G79950 helicase-related; K11136 regulator of telomere
elongation helicase 1
Length=1040
Score = 41.6 bits (96), Expect = 6e-04, Method: Composition-based stats.
Identities = 19/63 (30%), Positives = 35/63 (55%), Gaps = 1/63 (1%)
Query 32 IVARGADQVPLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYWY 91
+V+ G L S + R + N +++ + VP+GL+ FFPSY M++ +++W
Sbjct 525 VVSTGPSGYVLNSSYRNRDVPEYKQELGNAIVNFSRVVPEGLLIFFPSYYLMDSCITFW- 583
Query 92 ENG 94
+NG
Sbjct 584 KNG 586
> sce:YPL008W CHL1, CTF1, LPA9, MCM12; Chl1p (EC:3.6.1.-); K11273
chromosome transmission fidelity protein 1 [EC:3.6.4.13]
Length=861
Score = 41.2 bits (95), Expect = 9e-04, Method: Composition-based stats.
Identities = 23/63 (36%), Positives = 33/63 (52%), Gaps = 3/63 (4%)
Query 38 DQVPLTSKFECRHDVSVLRNYA-NLLLDLCKSVPD--GLVCFFPSYAYMETAMSYWYENG 94
+Q L FE R S++ N+ +DL K+VP G+V FFPSY Y+ + W +N
Sbjct 625 NQPELEFTFEKRMSPSLVNNHLFQFFVDLSKAVPKKGGIVAFFPSYQYLAHVIQCWKQND 684
Query 95 FLA 97
A
Sbjct 685 RFA 687
> tgo:TGME49_009770 DNA repair helicase, putative ; K11136 regulator
of telomere elongation helicase 1
Length=1649
Score = 41.2 bits (95), Expect = 0.001, Method: Composition-based stats.
Identities = 20/49 (40%), Positives = 27/49 (55%), Gaps = 0/49 (0%)
Query 42 LTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYW 90
L S F+ R+ LR LL +C V G++CFF SYA M T ++ W
Sbjct 797 LLSTFQNRNRPEYLRALGFSLLRICTLVGGGVLCFFASYAQMHTCITEW 845
> hsa:83990 BRIP1, BACH1, FANCJ, FLJ90232, MGC126521, MGC126523,
OF; BRCA1 interacting protein C-terminal helicase 1 (EC:3.6.4.13)
Length=1249
Score = 39.7 bits (91), Expect = 0.003, Method: Composition-based stats.
Identities = 18/53 (33%), Positives = 26/53 (49%), Gaps = 6/53 (11%)
Query 43 TSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGF 95
T FE + +V L LL +C++V G++CF PSY +E W G
Sbjct 667 TETFEFQDEVGAL------LLSVCQTVSQGILCFLPSYKLLEKLKERWLSTGL 713
> bbo:BBOV_III008820 17.m07769; DNA repair helicase (rad3) family
protein; K11136 regulator of telomere elongation helicase
1
Length=948
Score = 39.7 bits (91), Expect = 0.003, Method: Composition-based stats.
Identities = 20/71 (28%), Positives = 33/71 (46%), Gaps = 1/71 (1%)
Query 28 FCPMIVARG-ADQVPLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETA 86
CP ++ G D L+S + R S ++ +L + VP G++ FF SY +E
Sbjct 578 LCPRVITGGDVDGSILSSDYNNRSTPSYIKALGESVLTFVRCVPAGVLVFFVSYPVLEDT 637
Query 87 MSYWYENGFLA 97
++ W G A
Sbjct 638 VNAWKSAGIFA 648
> mmu:237911 Brip1, 3110009N10Rik, 8030460J03Rik, Bach1, FACJ,
Fancj, OF; BRCA1 interacting protein C-terminal helicase 1
(EC:3.6.4.13)
Length=1174
Score = 38.5 bits (88), Expect = 0.006, Method: Composition-based stats.
Identities = 18/53 (33%), Positives = 27/53 (50%), Gaps = 6/53 (11%)
Query 43 TSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGF 95
T FE + +V +L LL +C++V G++CF PSY +E W G
Sbjct 670 TETFEFQDEVGML------LLSVCQTVSQGILCFLPSYKLLEKLRERWIFTGL 716
> ath:AT1G20750 helicase-related
Length=1179
Score = 38.1 bits (87), Expect = 0.006, Method: Composition-based stats.
Identities = 17/54 (31%), Positives = 25/54 (46%), Gaps = 0/54 (0%)
Query 41 PLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENG 94
PL + + + L ++C VP G + FFPSY ME + W+E G
Sbjct 506 PLNASYRTAEAYAFQDALGKSLEEICTIVPGGSLVFFPSYKLMEKLCTRWHETG 559
> ath:AT1G20720 ATP binding / ATP-dependent DNA helicase/ ATP-dependent
helicase/ DNA binding / hydrolase, acting on acid
anhydrides, in phosphorus-containing anhydrides / nucleic acid
binding
Length=1175
Score = 37.4 bits (85), Expect = 0.012, Method: Composition-based stats.
Identities = 18/60 (30%), Positives = 26/60 (43%), Gaps = 0/60 (0%)
Query 33 VARGADQVPLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYWYE 92
++ G PL + ++ S L ++C VP G + FFPSY ME W E
Sbjct 527 ISNGPSNYPLNASYKTADAYSFQDALGKSLEEICTIVPGGSLVFFPSYKLMEKLCMRWRE 586
> pfa:PF14_0081 DNA-repair helicase, putative; K11136 regulator
of telomere elongation helicase 1
Length=1160
Score = 37.0 bits (84), Expect = 0.016, Method: Composition-based stats.
Identities = 15/49 (30%), Positives = 28/49 (57%), Gaps = 0/49 (0%)
Query 42 LTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYW 90
L S +E R + + +R+ N + D+ +P G++ FF SY+ M ++ W
Sbjct 613 LLSTYENRANENYIRSLGNCIFDIIVCIPYGVLIFFSSYSSMTETVNSW 661
> ath:AT1G79890 helicase-related; K11273 chromosome transmission
fidelity protein 1 [EC:3.6.4.13]
Length=882
Score = 35.8 bits (81), Expect = 0.030, Method: Composition-based stats.
Identities = 17/74 (22%), Positives = 33/74 (44%), Gaps = 0/74 (0%)
Query 30 PMIVARGADQVPLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSY 89
P+ V+ G R + +++ L+ +L VP+G++ FF S+ Y +
Sbjct 602 PIAVSHGPSGQSFDFSHSSRSSIGMIQELGLLMSNLVAVVPEGIIVFFSSFEYETQVHTA 661
Query 90 WYENGFLAQTLPFK 103
W +G L + + K
Sbjct 662 WSNSGILRRIVKKK 675
> cel:F25H2.13 rtel-1; RTEL (mammalian Regulator of TELomere length)
homolog family member (rtel-1); K11136 regulator of telomere
elongation helicase 1
Length=994
Score = 35.8 bits (81), Expect = 0.032, Method: Composition-based stats.
Identities = 18/59 (30%), Positives = 32/59 (54%), Gaps = 1/59 (1%)
Query 32 IVARGADQVPLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYW 90
IV RG + L F+ R ++ + A LL + + +P G++ FF SY+ M+ ++ W
Sbjct 527 IVTRG-KRGGLAGSFQNRKNLDYVTGVAEALLRVMEVIPQGILIFFSSYSQMDELVATW 584
> tpv:TP03_0644 DNA helicase; K11273 chromosome transmission fidelity
protein 1 [EC:3.6.4.13]
Length=740
Score = 35.8 bits (81), Expect = 0.038, Method: Composition-based stats.
Identities = 26/99 (26%), Positives = 48/99 (48%), Gaps = 11/99 (11%)
Query 1 PLELYPKILSFVPLIAESF-----PV-SMERACFCPMIVARGADQVPLTSKFECRHDVSV 54
P+E + LS P AE F PV ++R ++ + + L + R ++
Sbjct 461 PIE---EFLSLAPSRAEVFIHRSLPVFPLDR--LLSAVIHKDENGDELVFDYSRRENLKE 515
Query 55 LRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYWYEN 93
L+ L+ + + VP G+VCFF SYAY++ ++ ++
Sbjct 516 LKYLCTLMEIITEIVPGGIVCFFSSYAYLDVFYKFFLKS 554
> dre:794038 brip1, fancj, si:ch211-158l18.1; BRCA1 interacting
protein C-terminal helicase 1
Length=1217
Score = 35.0 bits (79), Expect = 0.052, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 61 LLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGF 95
LLL +C +V G++CF PSY ++ W G
Sbjct 708 LLLKVCHTVSRGVLCFLPSYKMLDKLRDRWTNTGL 742
> cel:M03C11.2 hypothetical protein; K11273 chromosome transmission
fidelity protein 1 [EC:3.6.4.13]
Length=848
Score = 34.7 bits (78), Expect = 0.066, Method: Composition-based stats.
Identities = 21/72 (29%), Positives = 32/72 (44%), Gaps = 0/72 (0%)
Query 33 VARGADQVPLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYWYE 92
V R D P ++ R + LR+ A + L +P+G+V F PSY ++ E
Sbjct 621 VERTVDGKPFQLTYQTRGADTTLRSLATSIQALIPHIPNGVVIFVPSYDFLFNFQKKMKE 680
Query 93 NGFLAQTLPFKA 104
G L + KA
Sbjct 681 FGILKRIEEKKA 692
> cpv:cgd7_3510 helicase ; K11273 chromosome transmission fidelity
protein 1 [EC:3.6.4.13]
Length=775
Score = 34.7 bits (78), Expect = 0.076, Method: Composition-based stats.
Identities = 21/89 (23%), Positives = 39/89 (43%), Gaps = 2/89 (2%)
Query 3 ELYPKILSFVPLIAESFPVS--MERACFCPMIVARGADQVPLTSKFECRHDVSVLRNYAN 60
E P +L P+ F S + +I+ + D + ++E R D+ L N
Sbjct 502 EFSPLLLPLEPMNIVKFSASHHISNNNLMCLIIPKFIDNSVIDLRYEFRTDIKQLLNLCE 561
Query 61 LLLDLCKSVPDGLVCFFPSYAYMETAMSY 89
LL L +P+G+ FF S+ +++ +
Sbjct 562 LLSVLSSEIPNGVCVFFTSHLFLDVFFKF 590
> bbo:BBOV_I004620 19.m02044; DNA repair helicase (rad3) and DEAD_2
domain containing protein; K11273 chromosome transmission
fidelity protein 1 [EC:3.6.4.13]
Length=775
Score = 34.3 bits (77), Expect = 0.091, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 33/56 (58%), Gaps = 3/56 (5%)
Query 35 RGADQVPLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYW 90
+G+ V ++ E + ++SVL NL+ L VP+G+VCF SY ++E+ S +
Sbjct 534 QGSTLVYDSNSREQKREISVL---FNLIERLSSVVPNGIVCFLSSYTFLESFQSAF 586
> tgo:TGME49_031800 hypothetical protein
Length=2272
Score = 32.3 bits (72), Expect = 0.40, Method: Composition-based stats.
Identities = 14/51 (27%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 40 VPLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYW 90
VPL + S L + ++ L +++P G++ FFPS++ ++ A W
Sbjct 1522 VPLECSSRQLTNPSFLLHLGWCIVRLVQAIPGGILVFFPSFSMLQKAQRLW 1572
> xla:398837 brip1, MGC68622; BRCA1 interacting protein C-terminal
helicase 1
Length=713
Score = 32.0 bits (71), Expect = 0.45, Method: Composition-based stats.
Identities = 11/20 (55%), Positives = 16/20 (80%), Gaps = 0/20 (0%)
Query 61 LLLDLCKSVPDGLVCFFPSY 80
LLL +C++V G++CF PSY
Sbjct 684 LLLSVCQTVSHGILCFLPSY 703
> cel:F33H2.1 dog-1; Deletions Of G-rich DNA family member (dog-1)
Length=983
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 11/34 (32%), Positives = 18/34 (52%), Gaps = 0/34 (0%)
Query 65 LCKSVPDGLVCFFPSYAYMETAMSYWYENGFLAQ 98
+C +VP G++CF PSY ++ N + Q
Sbjct 758 VCSNVPAGILCFLPSYRVLDQLKQCMIRNSTMRQ 791
> hsa:23325 KIAA1033
Length=1173
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 16/52 (30%), Positives = 24/52 (46%), Gaps = 10/52 (19%)
Query 53 SVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGFLAQTLPFKA 104
++ + + LLD+CK VP A TA W+ + FL Q +P A
Sbjct 331 TIDKKFYKSLLDICKKVP----------AITLTANIIWFPDNFLIQKIPAAA 372
> mmu:244650 Phlpp2, AI481772, C130044A18Rik, KIAA0931, Phlppl;
PH domain and leucine rich repeat protein phosphatase 2 (EC:3.1.3.16);
K01090 protein phosphatase [EC:3.1.3.16]
Length=1355
Score = 28.5 bits (62), Expect = 4.7, Method: Composition-based stats.
Identities = 11/34 (32%), Positives = 17/34 (50%), Gaps = 0/34 (0%)
Query 18 SFPVSMERACFCPMIVARGADQVPLTSKFECRHD 51
SF +++ C ++ RG VPL+ F HD
Sbjct 922 SFSLTVANVGMCQAVLCRGGKPVPLSKVFSLEHD 955
> xla:397974 mst1r, Xhlp, Xron; macrophage stimulating 1 receptor
(c-met-related tyrosine kinase); K05100 macrophage-stimulating
1 receptor [EC:2.7.10.1]
Length=1369
Score = 28.5 bits (62), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 32/73 (43%), Gaps = 6/73 (8%)
Query 37 ADQVPLT---SKFECRHDVSV-LRNYANLLLDLCKSV--PDGLVCFFPSYAYMETAMSYW 90
A PLT S + +SV L N + CK PD +VC P+Y Y+ + +
Sbjct 762 ASGTPLTIQGSNLDSASSISVILNNGEKAIRSACKGTFSPDRIVCRTPAYTYIGRSGNLT 821
Query 91 YENGFLAQTLPFK 103
E + T P++
Sbjct 822 LELDAVQYTYPYR 834
> tpv:TP02_0703 hypothetical protein
Length=3934
Score = 28.5 bits (62), Expect = 5.5, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 25/47 (53%), Gaps = 1/47 (2%)
Query 50 HDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGFL 96
H VS N N + L K + D +V +P+ Y++ S+WY+N L
Sbjct 1801 HKVSHALNNTNARMLLLK-MSDRVVRAYPTATYVKRVDSFWYKNSLL 1846
> tgo:TGME49_023950 hypothetical protein
Length=868
Score = 28.5 bits (62), Expect = 5.6, Method: Composition-based stats.
Identities = 17/55 (30%), Positives = 28/55 (50%), Gaps = 1/55 (1%)
Query 14 LIAESFP-VSMERACFCPMIVARGADQVPLTSKFECRHDVSVLRNYANLLLDLCK 67
L+ +S P + ++ F P G+ +T K CR + V+R +A LL+L K
Sbjct 324 LLTQSAPFLCLQEGPFSPHSGEAGSQTPRVTGKERCRRAMRVVRQWAAFLLELEK 378
> ath:AT2G34280 S locus F-box-related / SLF-related
Length=391
Score = 28.5 bits (62), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 11/30 (36%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 52 VSVLRNYANLLLDLCKSVPDGLVCFFPSYA 81
V +L + + L +C+S DGL+C + +YA
Sbjct 86 VRILTPWEDTLYKVCQSSCDGLICLYNTYA 115
> pfa:PF13_0144 oxidoreductase, putative (EC:1.1.1.-)
Length=334
Score = 28.1 bits (61), Expect = 7.1, Method: Composition-based stats.
Identities = 17/43 (39%), Positives = 22/43 (51%), Gaps = 3/43 (6%)
Query 47 ECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSY 89
E V RN LL+L +P+G VCF PS A +E +Y
Sbjct 216 EINQIVEKTRNTGFELLEL---LPEGSVCFAPSLAIVEIIEAY 255
Lambda K H
0.327 0.140 0.451
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2022291452
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40