bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_3381_orf1
Length=130
Score E
Sequences producing significant alignments: (Bits) Value
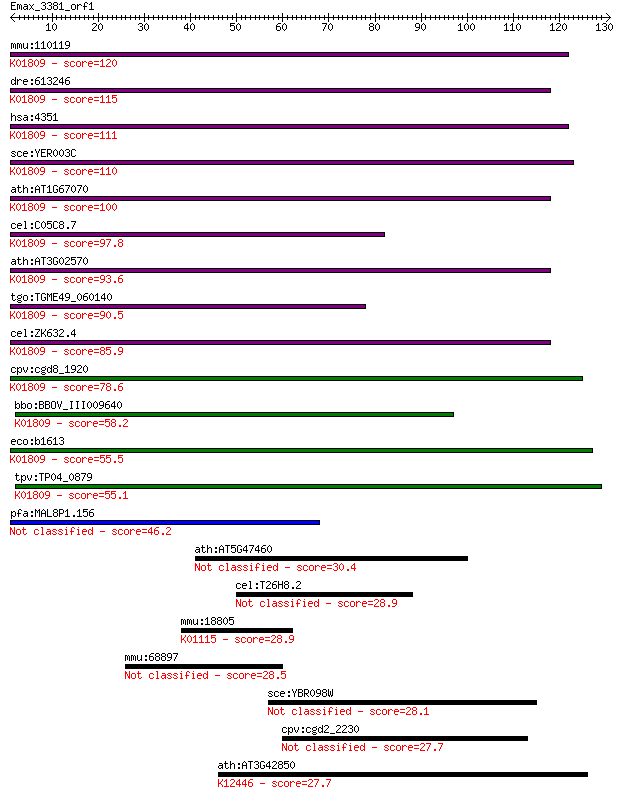
mmu:110119 Mpi, 1110002E17Rik, AI315153, Mpi-1, Mpi1; mannose ... 120 2e-27
dre:613246 mpi, MGC110773, zgc:110773; mannose phosphate isome... 115 4e-26
hsa:4351 MPI, CDG1B, FLJ39201, PMI, PMI1; mannose phosphate is... 111 5e-25
sce:YER003C PMI40; Mannose-6-phosphate isomerase, catalyzes th... 110 9e-25
ath:AT1G67070 DIN9; DIN9 (DARK INDUCIBLE 9); mannose-6-phospha... 100 1e-21
cel:C05C8.7 hypothetical protein; K01809 mannose-6-phosphate i... 97.8 8e-21
ath:AT3G02570 MEE31; MEE31 (MATERNAL EFFECT EMBRYO ARREST 31);... 93.6 1e-19
tgo:TGME49_060140 mannose-6-phosphate isomerase, putative (EC:... 90.5 1e-18
cel:ZK632.4 hypothetical protein; K01809 mannose-6-phosphate i... 85.9 3e-17
cpv:cgd8_1920 mannose-6-phosphate isomerase ; K01809 mannose-6... 78.6 5e-15
bbo:BBOV_III009640 17.m07836; phosphomannose isomerase type I ... 58.2 6e-09
eco:b1613 manA, ECK1608, JW1605, pmi; mannose-6-phosphate isom... 55.5 4e-08
tpv:TP04_0879 mannose-6-phosphate isomerase (EC:5.3.1.8); K018... 55.1 6e-08
pfa:MAL8P1.156 mannose-6-phosphate isomerase, putative (EC:5.3... 46.2 2e-05
ath:AT5G47460 pentatricopeptide (PPR) repeat-containing protein 30.4 1.4
cel:T26H8.2 srx-48; Serpentine Receptor, class X family member... 28.9 4.1
mmu:18805 Pld1, AA536939, C85393, Pld1a, Pld1b; phospholipase ... 28.9 4.4
mmu:68897 Disp1, 1190008H24Rik, DispA; dispatched homolog 1 (D... 28.5 5.2
sce:YBR098W MMS4, SLX2, YBR100W; Mms4p 28.1 6.4
cpv:cgd2_2230 hypothetical protein 27.7 9.0
ath:AT3G42850 galactokinase, putative; K12446 L-arabinokinase ... 27.7 9.7
> mmu:110119 Mpi, 1110002E17Rik, AI315153, Mpi-1, Mpi1; mannose
phosphate isomerase (EC:5.3.1.8); K01809 mannose-6-phosphate
isomerase [EC:5.3.1.8]
Length=423
Score = 120 bits (300), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 66/126 (52%), Positives = 81/126 (64%), Gaps = 5/126 (3%)
Query 1 FKVLSVRTALSIQAHPNKALAEKLHHDDPFHYPDANHKPELAIALTEFEAFCGFKPHAEI 60
FKVLSV TALSIQAHPNK LAEKLH P HYPDANHKPE+AIALT F+ CGF+P EI
Sbjct 98 FKVLSVDTALSIQAHPNKELAEKLHLQAPEHYPDANHKPEMAIALTSFQGLCGFRPVEEI 157
Query 61 EFLLNSVPGFKNIIGQDLVAKY-QSAGDKKE----TLKELFSALMTAPEELVKNNLSSLK 115
+ VP F+ +IG D A+ +S G E L+ FS LM + +++V L+ L
Sbjct 158 VTFMKKVPEFQLLIGDDATAQLKESVGGDTEAMASALRNCFSHLMKSEKKVVVEQLNLLV 217
Query 116 TRRTTQ 121
R + Q
Sbjct 218 KRISQQ 223
> dre:613246 mpi, MGC110773, zgc:110773; mannose phosphate isomerase
(EC:5.3.1.8); K01809 mannose-6-phosphate isomerase [EC:5.3.1.8]
Length=422
Score = 115 bits (287), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 61/121 (50%), Positives = 80/121 (66%), Gaps = 4/121 (3%)
Query 1 FKVLSVRTALSIQAHPNKALAEKLHHDDPFHYPDANHKPELAIALTEFEAFCGFKPHAEI 60
FKVLSV TALSIQAHPN+ LA +LH P HYPD NHKPE+AIALT+FE CGF+P EI
Sbjct 98 FKVLSVNTALSIQAHPNRELAARLHAQFPEHYPDNNHKPEMAIALTQFEGLCGFRPMEEI 157
Query 61 EFLLNSVPGFKNIIGQDLVAKYQSA-GDKKET---LKELFSALMTAPEELVKNNLSSLKT 116
L +VP F+ ++G + + QSA G+ T LK+ F+ +M ++L + L+ L
Sbjct 158 LGFLKNVPEFRALVGNEASEELQSADGNPHRTSLALKKCFTRMMNCEKKLFVDQLNMLVK 217
Query 117 R 117
R
Sbjct 218 R 218
> hsa:4351 MPI, CDG1B, FLJ39201, PMI, PMI1; mannose phosphate
isomerase (EC:5.3.1.8); K01809 mannose-6-phosphate isomerase
[EC:5.3.1.8]
Length=423
Score = 111 bits (278), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 63/126 (50%), Positives = 79/126 (62%), Gaps = 5/126 (3%)
Query 1 FKVLSVRTALSIQAHPNKALAEKLHHDDPFHYPDANHKPELAIALTEFEAFCGFKPHAEI 60
FKVLSV T LSIQAHPNK LAEKLH P HYPDANHKPE+AIALT F+ CGF+P EI
Sbjct 98 FKVLSVETPLSIQAHPNKELAEKLHLQAPQHYPDANHKPEMAIALTPFQGLCGFRPVEEI 157
Query 61 EFLLNSVPGFKNIIGQDLVA--KYQSAGDKK---ETLKELFSALMTAPEELVKNNLSSLK 115
L VP F+ +IG + K + D + +L+ FS LM + +++V L+ L
Sbjct 158 VTFLKKVPEFQFLIGDEAATHLKQTMSHDSQAVASSLQSCFSHLMKSEKKVVVEQLNLLV 217
Query 116 TRRTTQ 121
R + Q
Sbjct 218 KRISQQ 223
> sce:YER003C PMI40; Mannose-6-phosphate isomerase, catalyzes
the interconversion of fructose-6-P and mannose-6-P; required
for early steps in protein mannosylation (EC:5.3.1.8); K01809
mannose-6-phosphate isomerase [EC:5.3.1.8]
Length=429
Score = 110 bits (276), Expect = 9e-25, Method: Compositional matrix adjust.
Identities = 59/134 (44%), Positives = 80/134 (59%), Gaps = 12/134 (8%)
Query 1 FKVLSVRTALSIQAHPNKALAEKLHHDDPFHYPDANHKPELAIALTEFEAFCGFKPHAEI 60
FKVLS+ LSIQAHP+KAL + LH DP +YPD NHKPE+AIA+T+FE FCGFKP EI
Sbjct 97 FKVLSIEKVLSIQAHPDKALGKILHAQDPKNYPDDNHKPEMAIAVTDFEGFCGFKPLQEI 156
Query 61 EFLLNSVPGFKNIIGQ----DLVAKYQSAGDK--------KETLKELFSALMTAPEELVK 108
L +P +NI+G+ + + Q + K K+ L+ +FS +M A ++ +K
Sbjct 157 ADELKRIPELRNIVGEETSRNFIENIQPSAQKGSPEDEQNKKLLQAVFSRVMNASDDKIK 216
Query 109 NNLSSLKTRRTTQP 122
SL R P
Sbjct 217 IQARSLVERSKNSP 230
> ath:AT1G67070 DIN9; DIN9 (DARK INDUCIBLE 9); mannose-6-phosphate
isomerase (EC:5.3.1.8); K01809 mannose-6-phosphate isomerase
[EC:5.3.1.8]
Length=441
Score = 100 bits (248), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 54/123 (43%), Positives = 75/123 (60%), Gaps = 6/123 (4%)
Query 1 FKVLSVRTALSIQAHPNKALAEKLHHDDPFHYPDANHKPELAIALTEFEAFCGFKPHAEI 60
FKVLSV ALSIQAHPNKALAEKLH +DP Y D NHKPE+A+A+T F+A CGF E+
Sbjct 119 FKVLSVTKALSIQAHPNKALAEKLHREDPLLYRDNNHKPEIALAVTPFQALCGFVTLKEL 178
Query 61 EFLLNSVPGFKNIIGQDL------VAKYQSAGDKKETLKELFSALMTAPEELVKNNLSSL 114
+ ++ +VP ++G V ++ K ++ +F+ LM+A K +S +
Sbjct 179 KEVITNVPEITELVGSKAADQIFNVHEHDEDERIKSVVRLIFTQLMSASNNETKQVVSRM 238
Query 115 KTR 117
K R
Sbjct 239 KNR 241
> cel:C05C8.7 hypothetical protein; K01809 mannose-6-phosphate
isomerase [EC:5.3.1.8]
Length=467
Score = 97.8 bits (242), Expect = 8e-21, Method: Compositional matrix adjust.
Identities = 45/81 (55%), Positives = 53/81 (65%), Gaps = 0/81 (0%)
Query 1 FKVLSVRTALSIQAHPNKALAEKLHHDDPFHYPDANHKPELAIALTEFEAFCGFKPHAEI 60
KV+S+RT LS+Q HP K A +LH DP HYPD NHKPELA ALT FE CGF+P EI
Sbjct 89 MKVMSIRTTLSLQVHPTKEQARRLHEKDPIHYPDRNHKPELAYALTRFELLCGFRPAREI 148
Query 61 EFLLNSVPGFKNIIGQDLVAK 81
L + P F+ + G D AK
Sbjct 149 LQNLQTFPSFRLLFGGDTYAK 169
> ath:AT3G02570 MEE31; MEE31 (MATERNAL EFFECT EMBRYO ARREST 31);
mannose-6-phosphate isomerase (EC:5.3.1.8); K01809 mannose-6-phosphate
isomerase [EC:5.3.1.8]
Length=432
Score = 93.6 bits (231), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 50/122 (40%), Positives = 72/122 (59%), Gaps = 5/122 (4%)
Query 1 FKVLSVRTALSIQAHPNKALAEKLHHDDPFHYPDANHKPELAIALTEFEAFCGFKPHAEI 60
FKVLSV LSIQAHP+K LA+K+H P Y D NHKPE+A+A T+FEA CGF P E+
Sbjct 112 FKVLSVARPLSIQAHPDKKLAKKMHKAHPNLYKDDNHKPEMALAYTQFEALCGFIPLQEL 171
Query 61 EFLLNSVPGFKNIIG-----QDLVAKYQSAGDKKETLKELFSALMTAPEELVKNNLSSLK 115
+ ++ ++P + ++G Q K ++ +F+ LM+A + K +S LK
Sbjct 172 KSVIRAIPEIEELVGSEEANQVFCITEHDEEKVKSVVRTIFTLLMSADADTTKKIVSKLK 231
Query 116 TR 117
R
Sbjct 232 RR 233
> tgo:TGME49_060140 mannose-6-phosphate isomerase, putative (EC:5.3.1.8);
K01809 mannose-6-phosphate isomerase [EC:5.3.1.8]
Length=665
Score = 90.5 bits (223), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 42/77 (54%), Positives = 52/77 (67%), Gaps = 0/77 (0%)
Query 1 FKVLSVRTALSIQAHPNKALAEKLHHDDPFHYPDANHKPELAIALTEFEAFCGFKPHAEI 60
FKVLSV+ LSIQ+HP++ LAE LH P H+PD NHKPELAIA+ FEA CGF+ EI
Sbjct 213 FKVLSVKKPLSIQSHPDRQLAEILHRQHPQHFPDTNHKPELAIAVGPFEALCGFRAVEEI 272
Query 61 EFLLNSVPGFKNIIGQD 77
+ P K ++G D
Sbjct 273 VLFILHTPELKRVLGGD 289
> cel:ZK632.4 hypothetical protein; K01809 mannose-6-phosphate
isomerase [EC:5.3.1.8]
Length=416
Score = 85.9 bits (211), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 49/121 (40%), Positives = 70/121 (57%), Gaps = 4/121 (3%)
Query 1 FKVLSVRTALSIQAHPNKALAEKLHHDDPFHYPDANHKPELAIALTEFEAFCGFKPHAEI 60
FKVLSV LSIQ HP K + LH DP +YPD NHKPE+AIALTEFE GF+ H++I
Sbjct 87 FKVLSVLGPLSIQIHPTKEQGKLLHATDPKNYPDDNHKPEIAIALTEFELLSGFRQHSQI 146
Query 61 EFLLNSVPGFKNIIGQDLVAKYQSAGDKKET----LKELFSALMTAPEELVKNNLSSLKT 116
L + ++ ++ ++ S G E+ LK++FS + P+E ++ + L
Sbjct 147 SEYLKLYSEIQELLTEEEKSQIDSLGSYGESSAQVLKKIFSRIWRTPKEKLQIVVDKLAR 206
Query 117 R 117
R
Sbjct 207 R 207
> cpv:cgd8_1920 mannose-6-phosphate isomerase ; K01809 mannose-6-phosphate
isomerase [EC:5.3.1.8]
Length=576
Score = 78.6 bits (192), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 48/151 (31%), Positives = 76/151 (50%), Gaps = 27/151 (17%)
Query 1 FKVLSVRTALSIQAHPNKALAEKLHHDDPFHYPDANHKPELAIALTEFEAFCGFKPHAEI 60
K+LS+ LS+Q HP+K LA++LH P Y D NHKPE+AIAL+EFE FCGF+ EI
Sbjct 174 LKILSISKPLSLQVHPDKILAKELHSTFPCIYSDDNHKPEIAIALSEFETFCGFRNTIEI 233
Query 61 EFLLNSVPGFKNIIGQDL------VAKYQSAGDKKET---------------------LK 93
++ + P +I G L + +Y +K ++ K
Sbjct 234 LSIIENFPETFDIFGISLEFIKKTINEYNQQDNKLKSDVINVNNNIHDQSQEDPFFTIKK 293
Query 94 ELFSALMTAPEELVKNNLSSLKTRRTTQPET 124
+LF ++ + E + ++L+ L R + +T
Sbjct 294 DLFINMLNSKTETINDSLNKLVNRLKKKSQT 324
> bbo:BBOV_III009640 17.m07836; phosphomannose isomerase type
I family protein (EC:5.3.1.8); K01809 mannose-6-phosphate isomerase
[EC:5.3.1.8]
Length=464
Score = 58.2 bits (139), Expect = 6e-09, Method: Composition-based stats.
Identities = 30/95 (31%), Positives = 50/95 (52%), Gaps = 2/95 (2%)
Query 2 KVLSVRTALSIQAHPNKALAEKLHHDDPFHYPDANHKPELAIALTEFEAFCGFKPHAEIE 61
KVLS+ LS+Q HP+ A +L+ + D KPE+++AL F A CG +P +I+
Sbjct 129 KVLSIAKPLSLQLHPDPESALQLYMKNHPGISDNQAKPEMSVALCRFRALCGLRPLEQIK 188
Query 62 FLLNSVPGFKNIIGQDLVAKYQSAGDKKETLKELF 96
P F ++G D+++ + E + EL+
Sbjct 189 TYAQRYPPFAKMLGNDILSSISV--ETNENVGELY 221
> eco:b1613 manA, ECK1608, JW1605, pmi; mannose-6-phosphate isomerase
(EC:5.3.1.8); K01809 mannose-6-phosphate isomerase [EC:5.3.1.8]
Length=391
Score = 55.5 bits (132), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 49/136 (36%), Positives = 63/136 (46%), Gaps = 17/136 (12%)
Query 1 FKVLSVRTALSIQAHPNK-----ALAEKLHHDDPF-----HYPDANHKPELAIALTEFEA 50
FKVL LSIQ HPNK A++ P +Y D NHKPEL ALT F A
Sbjct 85 FKVLCAAQPLSIQVHPNKHNSEIGFAKENAAGIPMDAAERNYKDPNHKPELVFALTPFLA 144
Query 51 FCGFKPHAEIEFLLNSVPGFKNIIGQDLVAKYQSAGDKKETLKELFSALMTAPEELVKNN 110
F+ +EI LL V G +A + D E L ELF++L+ E
Sbjct 145 MNAFREFSEIVSLLQPVAG-----AHPAIAHFLQQPD-AERLSELFASLLNMQGEEKSRA 198
Query 111 LSSLKTRRTTQPETEP 126
L+ LK+ +Q + EP
Sbjct 199 LAILKSALDSQ-QGEP 213
> tpv:TP04_0879 mannose-6-phosphate isomerase (EC:5.3.1.8); K01809
mannose-6-phosphate isomerase [EC:5.3.1.8]
Length=454
Score = 55.1 bits (131), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 45/132 (34%), Positives = 65/132 (49%), Gaps = 15/132 (11%)
Query 2 KVLSVRTALSIQAHPNKALAEKLHHDDPFHYP---DANHKPELAIALTEFEAFCGFKPHA 58
KVLS++ LSIQ HP+ A +L YP D KPE+ IAL++F A CGF+
Sbjct 122 KVLSIQKPLSIQIHPDDKTAIELFTA---RYPGIVDNYSKPEMCIALSKFRAMCGFRDIF 178
Query 59 EIEFLLNSVPGFKNIIGQDLVAKYQSAGDKKETLKELFSALMTAPEELVKNNLSSL--KT 116
EI + FK +G++L + Y S + K E+ + + +P NL L K
Sbjct 179 EIFKFSDKHDEFKKFLGKELTSSY-SYSNPKTLYIEILNRVFFSP------NLGELAEKL 231
Query 117 RRTTQPETEPRL 128
R + T+P L
Sbjct 232 ARRLKGFTDPDL 243
> pfa:MAL8P1.156 mannose-6-phosphate isomerase, putative (EC:5.3.1.8)
Length=1011
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 23/67 (34%), Positives = 35/67 (52%), Gaps = 0/67 (0%)
Query 1 FKVLSVRTALSIQAHPNKALAEKLHHDDPFHYPDANHKPELAIALTEFEAFCGFKPHAEI 60
FK+LS+ LSIQ HPN+ L+ +P Y D K E+ + + CGF +I
Sbjct 265 FKMLSISKPLSIQIHPNEQQTLYLNTMNPLLYKDKIFKTEMCVCINSMTLLCGFMNIFKI 324
Query 61 EFLLNSV 67
FL+ ++
Sbjct 325 AFLIRNI 331
> ath:AT5G47460 pentatricopeptide (PPR) repeat-containing protein
Length=576
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 32/59 (54%), Gaps = 3/59 (5%)
Query 41 LAIALTEFEAFCGFKPHAEIEFLLNSVPGFKNIIGQDLVAKYQSAGDKKETLKELFSAL 99
+A AL + + CG HAE+ F ++P I+ ++++ Y GD E +K LF+ L
Sbjct 357 VASALIDMYSKCGMLKHAELMFW--TMPRKNLIVWNEMISGYARNGDSIEAIK-LFNQL 412
> cel:T26H8.2 srx-48; Serpentine Receptor, class X family member
(srx-48)
Length=303
Score = 28.9 bits (63), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 20/38 (52%), Gaps = 2/38 (5%)
Query 50 AFCGFKPHAEIEFLLNSVPGFKNIIGQDLVAKYQSAGD 87
AF GF + + FL+ +P KN G ++ QS GD
Sbjct 18 AFIGFMANWSVAFLIKKLPSLKNSFG--MLTTSQSIGD 53
> mmu:18805 Pld1, AA536939, C85393, Pld1a, Pld1b; phospholipase
D1 (EC:3.1.4.4); K01115 phospholipase D [EC:3.1.4.4]
Length=1036
Score = 28.9 bits (63), Expect = 4.4, Method: Composition-based stats.
Identities = 11/24 (45%), Positives = 15/24 (62%), Gaps = 0/24 (0%)
Query 38 KPELAIALTEFEAFCGFKPHAEIE 61
KPEL + +FCG + HAE+E
Sbjct 824 KPELGNKWINYISFCGLRTHAELE 847
> mmu:68897 Disp1, 1190008H24Rik, DispA; dispatched homolog 1
(Drosophila)
Length=1521
Score = 28.5 bits (62), Expect = 5.2, Method: Composition-based stats.
Identities = 13/40 (32%), Positives = 21/40 (52%), Gaps = 6/40 (15%)
Query 26 HDDPFHYPDANHK------PELAIALTEFEAFCGFKPHAE 59
H P+H+P ++H PE +A + A C +PH+E
Sbjct 82 HPCPYHHPVSSHSNHQECHPEAGLAASPALASCRMQPHSE 121
> sce:YBR098W MMS4, SLX2, YBR100W; Mms4p
Length=691
Score = 28.1 bits (61), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 37/69 (53%), Gaps = 13/69 (18%)
Query 57 HAEIEFLLNSVPGFKNIIGQ---DLVAKY--------QSAGDKKETLKELFSALMTAPEE 105
++ +EF+ NS+P ++IG+ D +Y +SA D ETLK+ F + P E
Sbjct 577 NSHMEFI-NSLPNLVSLIGKQRMDPAIRYMKYAHLNVKSAQDSTETLKKTFHQIGRMP-E 634
Query 106 LVKNNLSSL 114
+ NN+ SL
Sbjct 635 MKANNVVSL 643
> cpv:cgd2_2230 hypothetical protein
Length=833
Score = 27.7 bits (60), Expect = 9.0, Method: Composition-based stats.
Identities = 18/55 (32%), Positives = 32/55 (58%), Gaps = 2/55 (3%)
Query 60 IEFLLNSVPGFKNIIG--QDLVAKYQSAGDKKETLKELFSALMTAPEELVKNNLS 112
I+ LLN P +I+G +D + ++S DK + + L S++M + +L N+LS
Sbjct 561 IKDLLNHFPPQGDILGISKDEIPLFRSNSDKHKADRVLLSSVMNSKPKLQFNSLS 615
> ath:AT3G42850 galactokinase, putative; K12446 L-arabinokinase
[EC:2.7.1.46]
Length=964
Score = 27.7 bits (60), Expect = 9.7, Method: Composition-based stats.
Identities = 20/83 (24%), Positives = 34/83 (40%), Gaps = 5/83 (6%)
Query 46 TEFEAFCGFKPHAEIEFLLNSVPGFKNIIGQDLVAKYQSAGDKKETL-KELFSALMTAPE 104
T + C PH + +P ++I G++ + KY GD T+ K+ A+M
Sbjct 768 TSLDYLCNLSPHRFQALYASKLP--QSITGEEFLEKYGDHGDSVTTIDKDGTYAIMAPTR 825
Query 105 ELVKNN--LSSLKTRRTTQPETE 125
+ N + + K T P E
Sbjct 826 HPIYENFRVQAFKALLTATPSEE 848
Lambda K H
0.315 0.131 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2039374804
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40