bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_3332_orf1
Length=93
Score E
Sequences producing significant alignments: (Bits) Value
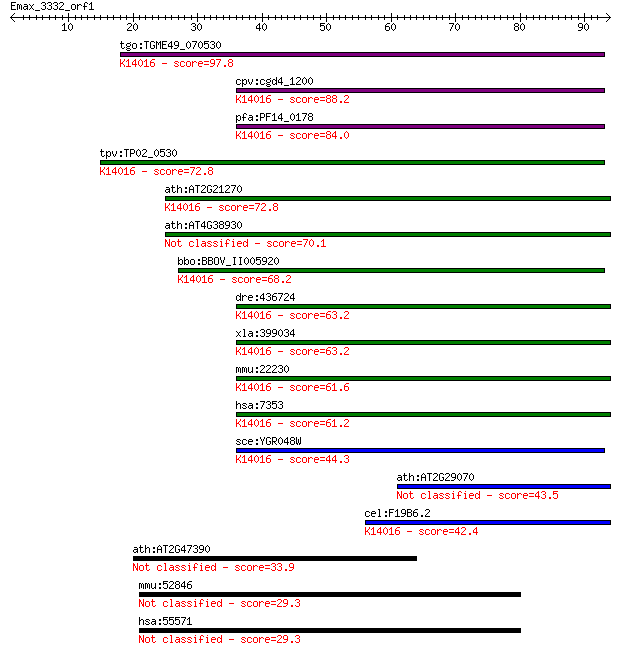
tgo:TGME49_070530 ubiquitin fusion degradation domain-containi... 97.8 7e-21
cpv:cgd4_1200 ubiquitin fusion degradation protein (UFD1); dou... 88.2 7e-18
pfa:PF14_0178 UFD1; Ubiquitin fusion degradation protein UFD1,... 84.0 1e-16
tpv:TP02_0530 hypothetical protein; K14016 ubiquitin fusion de... 72.8 3e-13
ath:AT2G21270 ubiquitin fusion degradation UFD1 family protein... 72.8 3e-13
ath:AT4G38930 ubiquitin fusion degradation UFD1 family protein 70.1 2e-12
bbo:BBOV_II005920 18.m06490; ubiquitin fusion degradation prot... 68.2 7e-12
dre:436724 ufd1l, wu:fc55f04, zgc:92341; ubiquitin fusion degr... 63.2 2e-10
xla:399034 ufd1l, MGC68571, ufd1; ubiquitin fusion degradation... 63.2 2e-10
mmu:22230 Ufd1l, Ufd1; ubiquitin fusion degradation 1 like; K1... 61.6 5e-10
hsa:7353 UFD1L, UFD1; ubiquitin fusion degradation 1 like (yea... 61.2 9e-10
sce:YGR048W UFD1; Protein that interacts with Cdc48p and Npl4p... 44.3 1e-04
ath:AT2G29070 ubiquitin fusion degradation UFD1 family protein 43.5 2e-04
cel:F19B6.2 ufd-1; Ubiquitin Fusion Degradation (yeast UFD hom... 42.4 4e-04
ath:AT2G47390 serine-type endopeptidase/ serine-type peptidase 33.9 0.13
mmu:52846 D1Bwg0212e, 2410015L18Rik, C40; DNA segment, Chr 1, ... 29.3 3.4
hsa:55571 C2orf29; chromosome 2 open reading frame 29 29.3
> tgo:TGME49_070530 ubiquitin fusion degradation domain-containing
protein (EC:3.1.3.48); K14016 ubiquitin fusion degradation
protein 1
Length=335
Score = 97.8 bits (242), Expect = 7e-21, Method: Composition-based stats.
Identities = 47/78 (60%), Positives = 54/78 (69%), Gaps = 3/78 (3%)
Query 18 LGRSMASFWGGGGEDGG---GFSASYTAYPVSFAAKDHLESGNKILLPPSALHALARLHI 74
R +A+ G G DGG GFS Y+ +PVSF KD +E GNKILLP SALHALARLHI
Sbjct 2 FSRHVANLLGMGEMDGGPGSGFSQCYSCFPVSFIGKDEMEKGNKILLPQSALHALARLHI 61
Query 75 SWPMHFRICNPAKDKLTH 92
SWPM F + N AKD+ TH
Sbjct 62 SWPMLFEVVNEAKDRRTH 79
> cpv:cgd4_1200 ubiquitin fusion degradation protein (UFD1); double
Psi beta barrel fold ; K14016 ubiquitin fusion degradation
protein 1
Length=322
Score = 88.2 bits (217), Expect = 7e-18, Method: Composition-based stats.
Identities = 38/57 (66%), Positives = 45/57 (78%), Gaps = 0/57 (0%)
Query 36 FSASYTAYPVSFAAKDHLESGNKILLPPSALHALARLHISWPMHFRICNPAKDKLTH 92
F Y+ YPVSFA +D LE GNKILLPPSAL+ LAR +I+WPM F+I NPAK+K TH
Sbjct 44 FINEYSCYPVSFAGRDELEGGNKILLPPSALNQLARRNITWPMLFQISNPAKNKFTH 100
> pfa:PF14_0178 UFD1; Ubiquitin fusion degradation protein UFD1,
putative; K14016 ubiquitin fusion degradation protein 1
Length=282
Score = 84.0 bits (206), Expect = 1e-16, Method: Composition-based stats.
Identities = 36/57 (63%), Positives = 42/57 (73%), Gaps = 0/57 (0%)
Query 36 FSASYTAYPVSFAAKDHLESGNKILLPPSALHALARLHISWPMHFRICNPAKDKLTH 92
F YT YPVSF KD +E+GNKI+LP +AL+ALAR HISWPM F + NP DK TH
Sbjct 22 FQEEYTCYPVSFIGKDDMENGNKIILPQTALNALARRHISWPMLFEVSNPYTDKRTH 78
> tpv:TP02_0530 hypothetical protein; K14016 ubiquitin fusion
degradation protein 1
Length=260
Score = 72.8 bits (177), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 36/79 (45%), Positives = 48/79 (60%), Gaps = 2/79 (2%)
Query 15 MWDLGRSMASFWGGGG-EDGGGFSASYTAYPVSFAAKDHLESGNKILLPPSALHALARLH 73
MW+ + +FW GG +++Y + VSFA ++ +E GNKILLP SALH LA +
Sbjct 1 MWNWN-NFENFWSGGNIYQNAAHTSNYRCFSVSFAGRESMEQGNKILLPQSALHELASRN 59
Query 74 ISWPMHFRICNPAKDKLTH 92
ISWPM F I NP K T+
Sbjct 60 ISWPMMFEILNPKNYKRTN 78
> ath:AT2G21270 ubiquitin fusion degradation UFD1 family protein;
K14016 ubiquitin fusion degradation protein 1
Length=319
Score = 72.8 bits (177), Expect = 3e-13, Method: Composition-based stats.
Identities = 34/69 (49%), Positives = 43/69 (62%), Gaps = 0/69 (0%)
Query 25 FWGGGGEDGGGFSASYTAYPVSFAAKDHLESGNKILLPPSALHALARLHISWPMHFRICN 84
F+ G G F SY YP SF K LESG+KI++PPSAL LA LHI +PM F + N
Sbjct 2 FFDGYHYHGTTFEQSYRCYPASFIDKPQLESGDKIIMPPSALDRLASLHIDYPMLFELRN 61
Query 85 PAKDKLTHC 93
+++THC
Sbjct 62 AGIERVTHC 70
> ath:AT4G38930 ubiquitin fusion degradation UFD1 family protein
Length=311
Score = 70.1 bits (170), Expect = 2e-12, Method: Composition-based stats.
Identities = 31/69 (44%), Positives = 41/69 (59%), Gaps = 0/69 (0%)
Query 25 FWGGGGEDGGGFSASYTAYPVSFAAKDHLESGNKILLPPSALHALARLHISWPMHFRICN 84
F+ G G F +Y YP SF K +ESG+KI++PPSAL LA L I +PM F + N
Sbjct 2 FYDGYAYHGTTFEQTYRCYPSSFIDKPQIESGDKIIMPPSALDRLASLQIDYPMLFELRN 61
Query 85 PAKDKLTHC 93
+ D +HC
Sbjct 62 ASTDSFSHC 70
> bbo:BBOV_II005920 18.m06490; ubiquitin fusion degradation protein;
K14016 ubiquitin fusion degradation protein 1
Length=258
Score = 68.2 bits (165), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 33/66 (50%), Positives = 41/66 (62%), Gaps = 0/66 (0%)
Query 27 GGGGEDGGGFSASYTAYPVSFAAKDHLESGNKILLPPSALHALARLHISWPMHFRICNPA 86
GG DG F Y YPVSF KD +ESGNKI +P SAL+ LA +I+WPM F + N
Sbjct 14 SGGYFDGLPFLVRYRCYPVSFLGKDAMESGNKICMPASALNELASRNITWPMMFELRNEE 73
Query 87 KDKLTH 92
+ + TH
Sbjct 74 RKRSTH 79
> dre:436724 ufd1l, wu:fc55f04, zgc:92341; ubiquitin fusion degradation
1-like; K14016 ubiquitin fusion degradation protein
1
Length=308
Score = 63.2 bits (152), Expect = 2e-10, Method: Composition-based stats.
Identities = 28/62 (45%), Positives = 40/62 (64%), Gaps = 4/62 (6%)
Query 36 FSASYTAYPVSFAA----KDHLESGNKILLPPSALHALARLHISWPMHFRICNPAKDKLT 91
FS Y Y VS A + +E G KI++PPSAL L+RL+I++PM F++ N D++T
Sbjct 19 FSTQYRCYSVSMLAGPNDRSDVEKGGKIIMPPSALDQLSRLNITYPMLFKLTNKNSDRMT 78
Query 92 HC 93
HC
Sbjct 79 HC 80
> xla:399034 ufd1l, MGC68571, ufd1; ubiquitin fusion degradation
1 like; K14016 ubiquitin fusion degradation protein 1
Length=307
Score = 63.2 bits (152), Expect = 2e-10, Method: Composition-based stats.
Identities = 28/62 (45%), Positives = 40/62 (64%), Gaps = 4/62 (6%)
Query 36 FSASYTAYPVSFAA----KDHLESGNKILLPPSALHALARLHISWPMHFRICNPAKDKLT 91
FS Y Y VS A + +E G KI++PPSAL L+RL+I++PM F++ N D++T
Sbjct 19 FSTQYRCYSVSMLAGPNDRSDVEKGGKIIMPPSALDQLSRLNITYPMLFKLTNKNSDRMT 78
Query 92 HC 93
HC
Sbjct 79 HC 80
> mmu:22230 Ufd1l, Ufd1; ubiquitin fusion degradation 1 like;
K14016 ubiquitin fusion degradation protein 1
Length=307
Score = 61.6 bits (148), Expect = 5e-10, Method: Composition-based stats.
Identities = 27/62 (43%), Positives = 40/62 (64%), Gaps = 4/62 (6%)
Query 36 FSASYTAYPVSFAA----KDHLESGNKILLPPSALHALARLHISWPMHFRICNPAKDKLT 91
FS Y + VS A + +E G KI++PPSAL L+RL+I++PM F++ N D++T
Sbjct 19 FSTQYRCFSVSMLAGPNDRSDVEKGGKIIMPPSALDQLSRLNITYPMLFKLTNKNSDRMT 78
Query 92 HC 93
HC
Sbjct 79 HC 80
> hsa:7353 UFD1L, UFD1; ubiquitin fusion degradation 1 like (yeast);
K14016 ubiquitin fusion degradation protein 1
Length=266
Score = 61.2 bits (147), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 27/62 (43%), Positives = 40/62 (64%), Gaps = 4/62 (6%)
Query 36 FSASYTAYPVSFAA----KDHLESGNKILLPPSALHALARLHISWPMHFRICNPAKDKLT 91
FS Y + VS A + +E G KI++PPSAL L+RL+I++PM F++ N D++T
Sbjct 19 FSTQYRCFSVSMLAGPNDRSDVEKGGKIIMPPSALDQLSRLNITYPMLFKLTNKNSDRMT 78
Query 92 HC 93
HC
Sbjct 79 HC 80
> sce:YGR048W UFD1; Protein that interacts with Cdc48p and Npl4p,
involved in recognition of polyubiquitinated proteins and
their presentation to the 26S proteasome for degradation;
involved in transporting proteins from the ER to the cytosol;
K14016 ubiquitin fusion degradation protein 1
Length=361
Score = 44.3 bits (103), Expect = 1e-04, Method: Composition-based stats.
Identities = 22/61 (36%), Positives = 32/61 (52%), Gaps = 4/61 (6%)
Query 36 FSASYTAYPVSFA----AKDHLESGNKILLPPSALHALARLHISWPMHFRICNPAKDKLT 91
F + YP++ KD G KI LPPSAL L+ L+I +PM F++ ++T
Sbjct 21 FEEFFRCYPIAMMNDRIRKDDANFGGKIFLPPSALSKLSMLNIRYPMLFKLTANETGRVT 80
Query 92 H 92
H
Sbjct 81 H 81
> ath:AT2G29070 ubiquitin fusion degradation UFD1 family protein
Length=280
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 17/33 (51%), Positives = 24/33 (72%), Gaps = 0/33 (0%)
Query 61 LPPSALHALARLHISWPMHFRICNPAKDKLTHC 93
+PPSAL LA LHI +PM F++ N + +K +HC
Sbjct 1 MPPSALDRLASLHIEYPMLFQLSNVSVEKTSHC 33
> cel:F19B6.2 ufd-1; Ubiquitin Fusion Degradation (yeast UFD homolog)
family member (ufd-1); K14016 ubiquitin fusion degradation
protein 1
Length=342
Score = 42.4 bits (98), Expect = 4e-04, Method: Composition-based stats.
Identities = 19/38 (50%), Positives = 26/38 (68%), Gaps = 0/38 (0%)
Query 56 GNKILLPPSALHALARLHISWPMHFRICNPAKDKLTHC 93
G KILLP SAL+ L + +I PM F++ N A ++THC
Sbjct 47 GGKILLPSSALNLLMQYNIPMPMLFKLTNMAVQRVTHC 84
> ath:AT2G47390 serine-type endopeptidase/ serine-type peptidase
Length=961
Score = 33.9 bits (76), Expect = 0.13, Method: Composition-based stats.
Identities = 18/44 (40%), Positives = 21/44 (47%), Gaps = 0/44 (0%)
Query 20 RSMASFWGGGGEDGGGFSASYTAYPVSFAAKDHLESGNKILLPP 63
RS+AS GG EDGGG S + + D L G LPP
Sbjct 71 RSLASACSGGAEDGGGTSNGSLSASATATEDDELAIGTGYRLPP 114
> mmu:52846 D1Bwg0212e, 2410015L18Rik, C40; DNA segment, Chr 1,
Brigham & Women's Genetics 0212 expressed
Length=505
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 20/66 (30%), Positives = 30/66 (45%), Gaps = 10/66 (15%)
Query 21 SMASFWGGGGEDGGGFSASYTAYPVSFAAKDHLESGNKIL-------LPPSALHALARLH 73
S+ S GGG G S ++ Y F+ DH G+ ++ L PSA L L+
Sbjct 68 SIISEEAGGGSTFEGLSTAFHHY---FSKADHFRLGSVLVMLLQQPDLLPSAAQRLTALY 124
Query 74 ISWPMH 79
+ W M+
Sbjct 125 LLWEMY 130
> hsa:55571 C2orf29; chromosome 2 open reading frame 29
Length=510
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 20/66 (30%), Positives = 30/66 (45%), Gaps = 10/66 (15%)
Query 21 SMASFWGGGGEDGGGFSASYTAYPVSFAAKDHLESGNKIL-------LPPSALHALARLH 73
S+ S GGG G S ++ Y F+ DH G+ ++ L PSA L L+
Sbjct 73 SIISEEAGGGSTFEGLSTAFHHY---FSKADHFRLGSVLVMLLQQPDLLPSAAQRLTALY 129
Query 74 ISWPMH 79
+ W M+
Sbjct 130 LLWEMY 135
Lambda K H
0.325 0.141 0.481
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2074765676
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40