bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_3331_orf2
Length=119
Score E
Sequences producing significant alignments: (Bits) Value
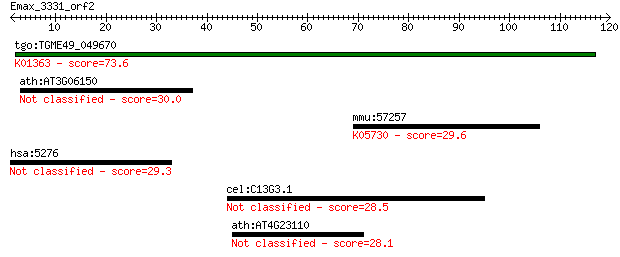
tgo:TGME49_049670 cysteine proteinase, putative (EC:3.4.22.1);... 73.6 2e-13
ath:AT3G06150 hypothetical protein 30.0 1.8
mmu:57257 Vav3, A530094I06Rik, AA986410, MGC27838; vav 3 oncog... 29.6 2.6
hsa:5276 SERPINI2, MEPI, PANCPIN, PI14, TSA2004; serpin peptid... 29.3 2.9
cel:C13G3.1 hypothetical protein 28.5 5.4
ath:AT4G23110 insulin-like growth factor binding 28.1 6.6
> tgo:TGME49_049670 cysteine proteinase, putative (EC:3.4.22.1);
K01363 cathepsin B [EC:3.4.22.1]
Length=572
Score = 73.6 bits (179), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 47/115 (40%), Positives = 63/115 (54%), Gaps = 6/115 (5%)
Query 2 NEAAEHSGANTPIHGPVLKTLAEGEFWSSRPAVSNAALRHLRLKMQRMKLSAAEKGNNPE 61
+E A+ +G + LK E FW SRPA SNAA+ ++ KM++ E +
Sbjct 166 SEQADEAGRDPGDLKKALKETGEDVFWESRPASSNAAVALIKRKMEKQAGKTGEGAGH-- 223
Query 62 LAVSWEAEVSPRFKYHSIKDAKRLMGTYLSWYEDPEKPAVPLGDPLPVKRFELQT 116
+WE EVS RF+Y S+KDAK+LMGT+L + P P G PLP K FE T
Sbjct 224 ---TWEPEVSLRFRYLSLKDAKKLMGTFLVNTKVEGFP-TPKGMPLPAKEFENAT 274
> ath:AT3G06150 hypothetical protein
Length=594
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 18/34 (52%), Gaps = 0/34 (0%)
Query 3 EAAEHSGANTPIHGPVLKTLAEGEFWSSRPAVSN 36
+A + SG N I G +T GE W SRP V +
Sbjct 174 QAIDESGKNVCIGGDYFETDISGENWKSRPPVKD 207
> mmu:57257 Vav3, A530094I06Rik, AA986410, MGC27838; vav 3 oncogene;
K05730 vav oncogene
Length=847
Score = 29.6 bits (65), Expect = 2.6, Method: Composition-based stats.
Identities = 14/38 (36%), Positives = 25/38 (65%), Gaps = 1/38 (2%)
Query 69 EVSPRFKYHSIKDAKRLMGTYLSW-YEDPEKPAVPLGD 105
E+ +K+HS+K+ R + T L + Y++PE+PA G+
Sbjct 741 ELVEYYKHHSLKEGFRTLDTTLQFPYKEPEQPAGQRGN 778
> hsa:5276 SERPINI2, MEPI, PANCPIN, PI14, TSA2004; serpin peptidase
inhibitor, clade I (pancpin), member 2
Length=405
Score = 29.3 bits (64), Expect = 2.9, Method: Composition-based stats.
Identities = 11/32 (34%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 1 VNEAAEHSGANTPIHGPVLKTLAEGEFWSSRP 32
+NE + +T IH PV+ +LA+ +F ++ P
Sbjct 339 INEDGSEAATSTGIHIPVIMSLAQSQFIANHP 370
> cel:C13G3.1 hypothetical protein
Length=136
Score = 28.5 bits (62), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 24/51 (47%), Gaps = 0/51 (0%)
Query 44 LKMQRMKLSAAEKGNNPELAVSWEAEVSPRFKYHSIKDAKRLMGTYLSWYE 94
L +Q + L+ KG+NP S V+ K K RL+ T +S ++
Sbjct 84 LPVQHLNLNLVRKGSNPSSQTSRNNSVTDLLKSELFKMPSRLLSTAVSLHQ 134
> ath:AT4G23110 insulin-like growth factor binding
Length=148
Score = 28.1 bits (61), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 45 KMQRMKLSAAEKGNNPELAVSWEAEV 70
K Q + +EK NNPE +SWE EV
Sbjct 119 KEQNKRRRISEKNNNPEETMSWEPEV 144
Lambda K H
0.312 0.129 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2022937320
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40