bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_3226_orf1
Length=116
Score E
Sequences producing significant alignments: (Bits) Value
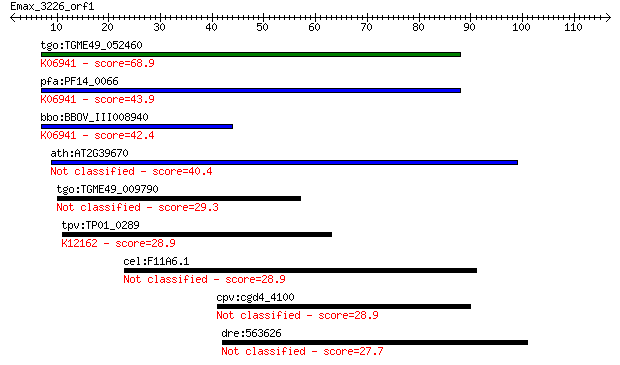
tgo:TGME49_052460 GPI transamidase 8, putative ; K06941 riboso... 68.9 4e-12
pfa:PF14_0066 radical SAM protein, putative; K06941 ribosomal ... 43.9 1e-04
bbo:BBOV_III008940 17.m07780; radical SAM domain containing pr... 42.4 4e-04
ath:AT2G39670 radical SAM domain-containing protein 40.4 0.002
tgo:TGME49_009790 radical SAM domain-containing protein (EC:1.... 29.3 3.2
tpv:TP01_0289 hypothetical protein; K12162 ubiquitin-fold modi... 28.9 3.9
cel:F11A6.1 kpc-1; Kex-2 Proprotein Convertase family member (... 28.9 4.1
cpv:cgd4_4100 hypothetical protein 28.9 4.1
dre:563626 fc59a02; wu:fc59a02 27.7 9.1
> tgo:TGME49_052460 GPI transamidase 8, putative ; K06941 ribosomal
RNA large subunit methyltransferase N [EC:2.1.1.-]
Length=270
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 36/81 (44%), Positives = 51/81 (62%), Gaps = 10/81 (12%)
Query 7 VPGVNDTVEHARALASLIRDRRRTTRHLYHVSLIEYNEASVSVDVMCAAQGVSSSLRRAD 66
+ G N+T EHA+ALA+L+R+RRR TRHLYHV++I YN AQGV SS++
Sbjct 157 IKGRNNTEEHAKALAALLRERRRPTRHLYHVNVIPYN----------TAQGVESSMQPPS 206
Query 67 SKRLKQFEEVLNGASISHSRR 87
+ + F ++L +S SRR
Sbjct 207 AAEVNHFTDLLRKLHLSVSRR 227
> pfa:PF14_0066 radical SAM protein, putative; K06941 ribosomal
RNA large subunit methyltransferase N [EC:2.1.1.-]
Length=362
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 27/82 (32%), Positives = 43/82 (52%), Gaps = 11/82 (13%)
Query 7 VPGVNDTVEHARALASLIRDRRRTTRHLYHVSLIEYNEASVSVDVMCAAQGVSSSLRRA- 65
+ +ND+ +HA AL+ I R R+LY+V LI YN+ A+ V + R
Sbjct 255 IKNLNDSKDHAEALSDHICKRPNNIRYLYNVCLIPYNK----------AKNVDENFHRLD 304
Query 66 DSKRLKQFEEVLNGASISHSRR 87
D++++ QFE++L IS R
Sbjct 305 DAEKILQFEKILKKNGISFFYR 326
> bbo:BBOV_III008940 17.m07780; radical SAM domain containing
protein; K06941 ribosomal RNA large subunit methyltransferase
N [EC:2.1.1.-]
Length=336
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 19/37 (51%), Positives = 24/37 (64%), Gaps = 0/37 (0%)
Query 7 VPGVNDTVEHARALASLIRDRRRTTRHLYHVSLIEYN 43
+ G NDT EHA L +I +R R+LYHV+LI YN
Sbjct 260 IKGENDTREHAEELVKVISNRPDEIRYLYHVNLIPYN 296
> ath:AT2G39670 radical SAM domain-containing protein
Length=428
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 38/90 (42%), Gaps = 16/90 (17%)
Query 9 GVNDTVEHARALASLIRDRRRTTRHLYHVSLIEYNEASVSVDVMCAAQGVSSSLRRADSK 68
GVND VEHA LA L+R+ +T YHV+LI YN S +R K
Sbjct 325 GVNDQVEHAVELAELLREWGKT----YHVNLIPYNPIE------------GSEYQRPYKK 368
Query 69 RLKQFEEVLNGASISHSRRWNHVADTTIMC 98
+ F L I+ S R D + C
Sbjct 369 AVLAFAAALESRKITASVRQTRGLDASAAC 398
> tgo:TGME49_009790 radical SAM domain-containing protein (EC:1.97.1.4)
Length=619
Score = 29.3 bits (64), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 23/47 (48%), Gaps = 4/47 (8%)
Query 10 VNDTVEHARALASLIRDRRRTTRHLYHVSLIEYNEASVSVDVMCAAQ 56
VNDT+E A AL +++ R V+LI YN V D + Q
Sbjct 384 VNDTLEQAHALGRILQPR----ADAVIVNLIPYNPTDVPYDYKPSTQ 426
> tpv:TP01_0289 hypothetical protein; K12162 ubiquitin-fold modifier
1
Length=998
Score = 28.9 bits (63), Expect = 3.9, Method: Composition-based stats.
Identities = 14/52 (26%), Positives = 28/52 (53%), Gaps = 0/52 (0%)
Query 11 NDTVEHARALASLIRDRRRTTRHLYHVSLIEYNEASVSVDVMCAAQGVSSSL 62
+D + H +AL S + + + H+SL EY+E+ + VD+ + +S +
Sbjct 919 SDYISHIKALVSSVETLNSSLTNPKHISLSEYSESFLIVDIEMLKKQISRDI 970
> cel:F11A6.1 kpc-1; Kex-2 Proprotein Convertase family member
(kpc-1)
Length=760
Score = 28.9 bits (63), Expect = 4.1, Method: Composition-based stats.
Identities = 21/75 (28%), Positives = 32/75 (42%), Gaps = 7/75 (9%)
Query 23 LIRDRRRTTRHLYHVSLIEYNEASVSVDVMCAAQGVSSSLRRADSKRLKQ-------FEE 75
L RD R+ +R + N+ DVM Q V+ + +R+++ FEE
Sbjct 97 LFRDDRKKSRSSRKTRSLSANQLQHEEDVMWMEQQVAKRRVKRGYRRIRRHTDDNDIFEE 156
Query 76 VLNGASISHSRRWNH 90
+G IS SR H
Sbjct 157 DDDGTQISKSRNRKH 171
> cpv:cgd4_4100 hypothetical protein
Length=312
Score = 28.9 bits (63), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 25/51 (49%), Gaps = 2/51 (3%)
Query 41 EYNEASVSVDVMCAAQGVSSSLRRADSKRLKQFEEVLNGASISHSRR--WN 89
EYN + ++ M G +SS R R Q +LNG S+ H +R WN
Sbjct 195 EYNSSFITYANMQTDFGENSSFRLPSKARDSQSIIILNGISVIHMQRQHWN 245
> dre:563626 fc59a02; wu:fc59a02
Length=823
Score = 27.7 bits (60), Expect = 9.1, Method: Composition-based stats.
Identities = 18/59 (30%), Positives = 29/59 (49%), Gaps = 2/59 (3%)
Query 42 YNEASVSVDVMCAAQGVSSSLRRADSKRLKQFEEVLNGASISHSRRWNHVADTTIMCKL 100
Y + + +D + +AQ +S + L F + N IS++RRWN V T I +L
Sbjct 747 YQDVLLQMDFIHSAQFLSRLPENTPAHTL--FSCIANTQMISNNRRWNQVFSTLIKDRL 803
Lambda K H
0.320 0.127 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2037741960
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40