bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_3190_orf1
Length=116
Score E
Sequences producing significant alignments: (Bits) Value
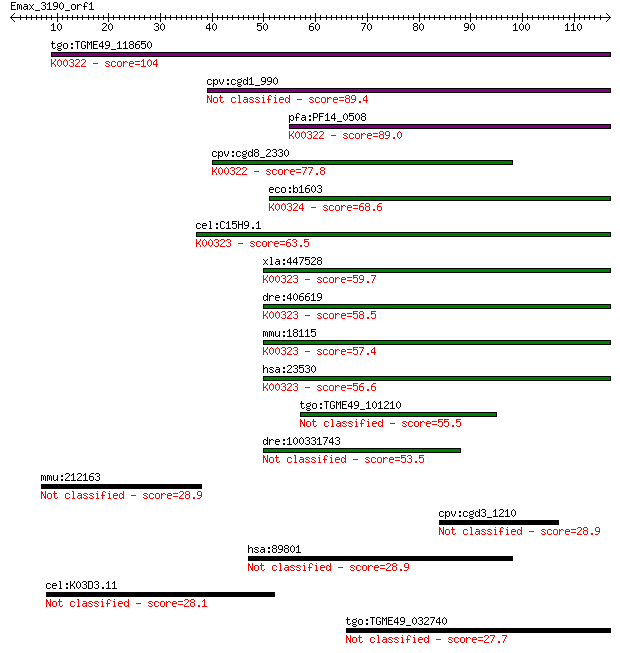
tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00... 104 6e-23
cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alph... 89.4 2e-18
pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative (... 89.0 3e-18
cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alp... 77.8 8e-15
eco:b1603 pntA, ECK1598, JW1595; pyridine nucleotide transhydr... 68.6 5e-12
cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase fa... 63.5 2e-10
xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydroge... 59.7 2e-09
dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamid... 58.5 5e-09
mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide... 57.4 1e-08
hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide t... 56.6 2e-08
tgo:TGME49_101210 NAD(P) transhydrogenase, alpha subunit, puta... 55.5 4e-08
dre:100331743 Nicotinamide Nucleotide Transhydrogenase family ... 53.5 2e-07
mmu:212163 8030462N17Rik, Akd2; RIKEN cDNA 8030462N17 gene 28.9 4.2
cpv:cgd3_1210 hypothetical protein 28.9 4.3
hsa:89801 PPP1R3F, HB2E; protein phosphatase 1, regulatory (in... 28.9 4.5
cel:K03D3.11 srz-104; Serpentine Receptor, class Z family memb... 28.1 6.9
tgo:TGME49_032740 hypothetical protein 27.7 9.6
> tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00322
NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1013
Score = 104 bits (260), Expect = 6e-23, Method: Compositional matrix adjust.
Identities = 58/108 (53%), Positives = 71/108 (65%), Gaps = 1/108 (0%)
Query 9 PNVAALVHSDQSSDYFVVHVDTEVAAAVVGLLGIVLDPMEPKHLTPLALSLIVGYYCVWA 68
P +L+ +SD F + AA GLLG+ L +E + T LALSLIVGYY VW+
Sbjct 893 PKEVSLLDQLVTSDAFFA-MSLVCTAAFAGLLGVTLKALELQQFTLLALSLIVGYYSVWS 951
Query 69 VTPALHTPLMSVTNALSGVIVIGCMLEYGTALISGFTLLAIIGTFLAS 116
VTPALHTPLMSVTNALSGVI+IG MLEYG + S + A+ TF +S
Sbjct 952 VTPALHTPLMSVTNALSGVIIIGSMLEYGPSATSASAICALCATFFSS 999
> cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, 12 transmembrane domain
(EC:1.6.1.2)
Length=1147
Score = 89.4 bits (220), Expect = 2e-18, Method: Composition-based stats.
Identities = 39/78 (50%), Positives = 58/78 (74%), Gaps = 0/78 (0%)
Query 39 LLGIVLDPMEPKHLTPLALSLIVGYYCVWAVTPALHTPLMSVTNALSGVIVIGCMLEYGT 98
+LG+ + ++ +++ +S ++GYYCVW V P LHTPLMSVTNALSGVI+IG M++YG
Sbjct 1051 VLGLTMTTIQIQNIFSFIISTMLGYYCVWDVDPKLHTPLMSVTNALSGVIIIGSMMQYGN 1110
Query 99 ALISGFTLLAIIGTFLAS 116
++ TL+++ TFLAS
Sbjct 1111 QTVTYTTLMSMFSTFLAS 1128
> pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative
(EC:1.6.1.2); K00322 NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1176
Score = 89.0 bits (219), Expect = 3e-18, Method: Composition-based stats.
Identities = 40/62 (64%), Positives = 50/62 (80%), Gaps = 0/62 (0%)
Query 55 LALSLIVGYYCVWAVTPALHTPLMSVTNALSGVIVIGCMLEYGTALISGFTLLAIIGTFL 114
LS IVGYYCVW+VTPALHTPLMS+TNALSGVI+IG M+EYG ++L+++ TFL
Sbjct 1096 FTLSTIVGYYCVWSVTPALHTPLMSMTNALSGVIIIGSMIEYGNGYKDLSSILSMLATFL 1155
Query 115 AS 116
+S
Sbjct 1156 SS 1157
> cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, possible signal peptide
plus 12 transmembrane regions ; K00322 NAD(P) transhydrogenase
[EC:1.6.1.1]
Length=1143
Score = 77.8 bits (190), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 36/58 (62%), Positives = 46/58 (79%), Gaps = 0/58 (0%)
Query 40 LGIVLDPMEPKHLTPLALSLIVGYYCVWAVTPALHTPLMSVTNALSGVIVIGCMLEYG 97
LG ++D ++ +LS+IVGYYC+W VTP+LHTPLMSVTNALSG+I+IG MLE G
Sbjct 1024 LGYIIDHDTLGNILVFSLSVIVGYYCIWNVTPSLHTPLMSVTNALSGIIIIGAMLECG 1081
> eco:b1603 pntA, ECK1598, JW1595; pyridine nucleotide transhydrogenase,
alpha subunit (EC:1.6.1.2); K00324 NAD(P) transhydrogenase
subunit alpha [EC:1.6.1.2]
Length=510
Score = 68.6 bits (166), Expect = 5e-12, Method: Composition-based stats.
Identities = 34/66 (51%), Positives = 45/66 (68%), Gaps = 2/66 (3%)
Query 51 HLTPLALSLIVGYYCVWAVTPALHTPLMSVTNALSGVIVIGCMLEYGTALISGFTLLAII 110
H T AL+ +VGYY VW V+ ALHTPLMSVTNA+SG+IV+G +L+ G + L+ I
Sbjct 427 HFTVFALACVVGYYVVWNVSHALHTPLMSVTNAISGIIVVGALLQIGQG--GWVSFLSFI 484
Query 111 GTFLAS 116
+AS
Sbjct 485 AVLIAS 490
> cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase
family member (nnt-1); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1041
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 37/83 (44%), Positives = 51/83 (61%), Gaps = 3/83 (3%)
Query 37 VGLLGIV-LDPMEPKHLTPLALSLIVGYYCVWAVTPALHTPLMSVTNALSGVIVIG--CM 93
V LLGI +P T AL+ +VGY+ VW VTPALH+PLMSVTNA+SG G C+
Sbjct 440 VSLLGIAGTNPQISSMSTTFALAGLVGYHTVWGVTPALHSPLMSVTNAISGTTAAGALCL 499
Query 94 LEYGTALISGFTLLAIIGTFLAS 116
+ G + +A++ TF++S
Sbjct 500 MGGGLMPQNSAQTMALLATFISS 522
> xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydrogenase
(EC:1.6.1.2); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1086
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 32/69 (46%), Positives = 46/69 (66%), Gaps = 2/69 (2%)
Query 50 KHLTPLALSLIVGYYCVWAVTPALHTPLMSVTNALSGVIVIGCMLEYGTALISGFT--LL 107
+ +T L+ IVGY+ VW VTPALH+PLMSVTNA+SG+ +G + G + T LL
Sbjct 500 QMVTTFGLAGIVGYHTVWGVTPALHSPLMSVTNAISGLTAVGGLALMGGGYLPTNTHELL 559
Query 108 AIIGTFLAS 116
A++ F++S
Sbjct 560 AVLAAFVSS 568
> dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P)
transhydrogenase [EC:1.6.1.2]
Length=1079
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 31/69 (44%), Positives = 45/69 (65%), Gaps = 2/69 (2%)
Query 50 KHLTPLALSLIVGYYCVWAVTPALHTPLMSVTNALSGVIVIGCMLEYGTALISGFT--LL 107
+ +T L+ IVGY+ VW VTPALH+PLMSVTNA+SG+ +G + G + T L
Sbjct 496 QMVTTFGLAGIVGYHTVWGVTPALHSPLMSVTNAISGLTAVGGLSLMGGGYLPSSTAETL 555
Query 108 AIIGTFLAS 116
A++ F++S
Sbjct 556 AVLAAFISS 564
> mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 33/74 (44%), Positives = 46/74 (62%), Gaps = 12/74 (16%)
Query 50 KHLTPLALSLIVGYYCVWAVTPALHTPLMSVTNALSGVIVIGCMLEYGTALISGF----- 104
+ +T L+ I+GY+ VW VTPALH+PLMSVTNA+SG+ +G G AL+ G
Sbjct 500 QMVTTFGLAGIIGYHTVWGVTPALHSPLMSVTNAISGLTAVG-----GLALMGGHFYPST 554
Query 105 --TLLAIIGTFLAS 116
LA + TF++S
Sbjct 555 TSQSLAALATFISS 568
> hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide
transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 32/69 (46%), Positives = 44/69 (63%), Gaps = 2/69 (2%)
Query 50 KHLTPLALSLIVGYYCVWAVTPALHTPLMSVTNALSGVIVIGCMLEYGTALISGFTL--L 107
+ +T L+ IVGY+ VW VTPALH+PLMSVTNA+SG+ +G + G L T L
Sbjct 500 QMVTTFGLAGIVGYHTVWGVTPALHSPLMSVTNAISGLTAVGGLALMGGHLYPSTTSQGL 559
Query 108 AIIGTFLAS 116
A + F++S
Sbjct 560 AALAAFISS 568
> tgo:TGME49_101210 NAD(P) transhydrogenase, alpha subunit, putative
(EC:1.6.1.2)
Length=1165
Score = 55.5 bits (132), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 25/38 (65%), Positives = 30/38 (78%), Gaps = 0/38 (0%)
Query 57 LSLIVGYYCVWAVTPALHTPLMSVTNALSGVIVIGCML 94
L+ VGY VW V PALHTPLMSVTNA+SG +++G ML
Sbjct 1057 LACFVGYLLVWNVAPALHTPLMSVTNAISGTVLVGGML 1094
> dre:100331743 Nicotinamide Nucleotide Transhydrogenase family
member (nnt-1)-like
Length=518
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 24/38 (63%), Positives = 31/38 (81%), Gaps = 0/38 (0%)
Query 50 KHLTPLALSLIVGYYCVWAVTPALHTPLMSVTNALSGV 87
+ +T L+ IVGY+ VW VTPALH+PLMSVTNA+SG+
Sbjct 200 QMVTTFGLAGIVGYHTVWGVTPALHSPLMSVTNAISGL 237
> mmu:212163 8030462N17Rik, Akd2; RIKEN cDNA 8030462N17 gene
Length=399
Score = 28.9 bits (63), Expect = 4.2, Method: Composition-based stats.
Identities = 17/31 (54%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 7 GLPNVAALVHSDQSSDYFVVHVDTEVAAAVV 37
G P VA SD SSD V++VD +AAAVV
Sbjct 240 GPPVVAHYDMSDTSSDPEVINVDNLLAAAVV 270
> cpv:cgd3_1210 hypothetical protein
Length=464
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 12/23 (52%), Positives = 17/23 (73%), Gaps = 0/23 (0%)
Query 84 LSGVIVIGCMLEYGTALISGFTL 106
LS +I+I +L +G+AL SG TL
Sbjct 9 LSALIIITLLLSFGSALFSGLTL 31
> hsa:89801 PPP1R3F, HB2E; protein phosphatase 1, regulatory (inhibitor)
subunit 3F
Length=453
Score = 28.9 bits (63), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 25/51 (49%), Gaps = 3/51 (5%)
Query 47 MEPKHLTPLALSLIVGYYCVWAVTPALHTPLMSVTNALSGVIVIGCMLEYG 97
+EP +P +L V CV A+ P L PL L+G++V+ L G
Sbjct 390 LEPPKKSP---TLAVPAECVCALPPQLRGPLTQTLGVLAGLVVVPVALNSG 437
> cel:K03D3.11 srz-104; Serpentine Receptor, class Z family member
(srz-104)
Length=261
Score = 28.1 bits (61), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 13/44 (29%), Positives = 25/44 (56%), Gaps = 1/44 (2%)
Query 8 LPNVAALVHSDQSSDYFVVHVDTEVAAAVVGLLGIVLDPMEPKH 51
LP V L + Q+S Y V+++ + + +VG++ + D M+ H
Sbjct 6 LPIVTHLYKTGQNSSYLVIYISISILSYLVGMVWNIFD-MDQDH 48
> tgo:TGME49_032740 hypothetical protein
Length=380
Score = 27.7 bits (60), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 26/51 (50%), Gaps = 0/51 (0%)
Query 66 VWAVTPALHTPLMSVTNALSGVIVIGCMLEYGTALISGFTLLAIIGTFLAS 116
+W + P P+++ N L V+ M E+ TA +S + +IG+F A
Sbjct 200 LWILRPLPCLPMLTGANCLGSVLYPITMSEFTTADVSDSVMNGVIGSFPAK 250
Lambda K H
0.324 0.140 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2037741960
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40