bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_3172_orf2
Length=129
Score E
Sequences producing significant alignments: (Bits) Value
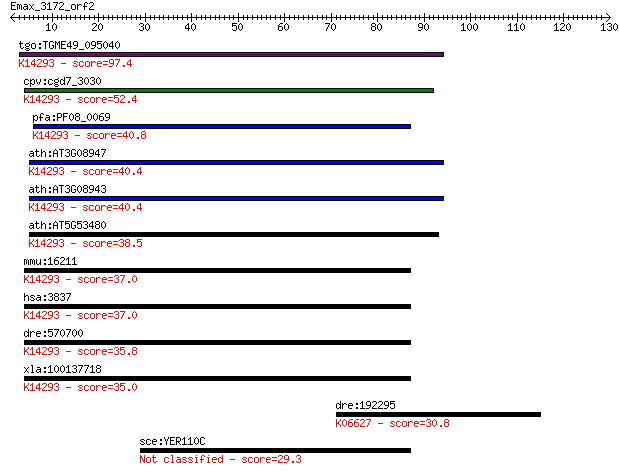
tgo:TGME49_095040 importin subunit beta-1, putative ; K14293 i... 97.4 1e-20
cpv:cgd7_3030 importin/karyopherin ; K14293 importin subunit b... 52.4 3e-07
pfa:PF08_0069 importin beta, putative; K14293 importin subunit... 40.8 0.001
ath:AT3G08947 binding / protein transporter; K14293 importin s... 40.4 0.001
ath:AT3G08943 binding / protein transporter; K14293 importin s... 40.4 0.001
ath:AT5G53480 importin beta-2, putative; K14293 importin subun... 38.5 0.005
mmu:16211 Kpnb1, AA409963, IPOB, Impnb, MGC8315; karyopherin (... 37.0 0.015
hsa:3837 KPNB1, IMB1, IPO1, IPOB, Impnb, MGC2155, MGC2156, MGC... 37.0 0.016
dre:570700 kpnb1, MGC123184, wu:fc12b10, zgc:123184; karyopher... 35.8 0.034
xla:100137718 kpnb1, imb1, impnb, ipo1, ipob, ntf97; karyopher... 35.0 0.066
dre:192295 ccna2, CycA2, cb671, chunp6924, wu:fi36f03; cyclin ... 30.8 1.2
sce:YER110C KAP123, YRB4; Kap123p 29.3 3.5
> tgo:TGME49_095040 importin subunit beta-1, putative ; K14293
importin subunit beta-1
Length=971
Score = 97.4 bits (241), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 53/91 (58%), Positives = 65/91 (71%), Gaps = 0/91 (0%)
Query 3 LIAKQIAAVTFKNSISAKDMTLDGAAAQAWRALPAPTKAEIKKQLLAALQTECTQVGNAV 62
L+AKQIAAVTFKN ISAKD+ LD AAA WRA+ K ++ QLLAA++TE QV NAV
Sbjct 74 LLAKQIAAVTFKNCISAKDVVLDSAAADKWRAVAEAAKQAMRLQLLAAIKTEHIQVANAV 133
Query 63 CQALAKLGVIELRSLEFEDLLPALQDMVQQA 93
CQ L+K+G IEL F +LLP L +V +A
Sbjct 134 CQVLSKIGRIELPGDGFPELLPFLLTLVTEA 164
> cpv:cgd7_3030 importin/karyopherin ; K14293 importin subunit
beta-1
Length=882
Score = 52.4 bits (124), Expect = 3e-07, Method: Composition-based stats.
Identities = 28/88 (31%), Positives = 49/88 (55%), Gaps = 0/88 (0%)
Query 4 IAKQIAAVTFKNSISAKDMTLDGAAAQAWRALPAPTKAEIKKQLLAALQTECTQVGNAVC 63
+++Q+A + KN++S + +D W +LP ++IK +L ++ + V A C
Sbjct 52 LSRQLAGLLLKNAVSGIEPRIDIERRGMWISLPQNVTSKIKALVLESILSPVASVRGASC 111
Query 64 QALAKLGVIELRSLEFEDLLPALQDMVQ 91
Q +AKLG +EL + +LLP L +VQ
Sbjct 112 QVIAKLGRVELPCKRWPELLPYLIRLVQ 139
> pfa:PF08_0069 importin beta, putative; K14293 importin subunit
beta-1
Length=877
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 27/83 (32%), Positives = 42/83 (50%), Gaps = 3/83 (3%)
Query 6 KQIAAVTFKNSISAKDMTLDGAAAQAWRALPAPTKAEIKKQLLAAL--QTECTQVGNAVC 63
+QIA + KN+ ++KD A+ W P K E+K LL L Q++ +G A C
Sbjct 57 RQIAGLLIKNAFTSKDNYESEEKARTWLNFPEDIKMELKNNLLVLLSQQSDKIVIGTA-C 115
Query 64 QALAKLGVIELRSLEFEDLLPAL 86
Q ++ + IEL + +LL L
Sbjct 116 QIISIIAKIELSHNKSSELLHKL 138
> ath:AT3G08947 binding / protein transporter; K14293 importin
subunit beta-1
Length=873
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/90 (27%), Positives = 48/90 (53%), Gaps = 1/90 (1%)
Query 5 AKQIAAVTFKNSISAKDMTLDGAAAQAWRALPAPTKAEIKKQLLAALQTECTQVGNAVCQ 64
++++A + KNS+ AKD + W A+ K++IK +LL L + + + Q
Sbjct 55 SRRLAGILLKNSLDAKDSATKDHLVKQWFAIDVALKSQIKDRLLRTLGSSALEARHTSAQ 114
Query 65 ALAKLGVIELRSLEFEDLLPA-LQDMVQQA 93
+AK+ IE+ ++ +L+ + L +M QQ
Sbjct 115 VIAKVASIEIPQKQWPELVGSLLNNMTQQG 144
> ath:AT3G08943 binding / protein transporter; K14293 importin
subunit beta-1
Length=871
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/90 (27%), Positives = 48/90 (53%), Gaps = 1/90 (1%)
Query 5 AKQIAAVTFKNSISAKDMTLDGAAAQAWRALPAPTKAEIKKQLLAALQTECTQVGNAVCQ 64
++++A + KNS+ AKD + W A+ K++IK +LL L + + + Q
Sbjct 55 SRRLAGILLKNSLDAKDSATKDHLVKQWFAIDVALKSQIKDRLLRTLGSSALEARHTSAQ 114
Query 65 ALAKLGVIELRSLEFEDLLPA-LQDMVQQA 93
+AK+ IE+ ++ +L+ + L +M QQ
Sbjct 115 VIAKVASIEIPQKQWPELVGSLLNNMTQQG 144
> ath:AT5G53480 importin beta-2, putative; K14293 importin subunit
beta-1
Length=870
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 25/88 (28%), Positives = 45/88 (51%), Gaps = 0/88 (0%)
Query 5 AKQIAAVTFKNSISAKDMTLDGAAAQAWRALPAPTKAEIKKQLLAALQTECTQVGNAVCQ 64
++++A + KN++ AK+ Q W AL TK++I+ LL L V + Q
Sbjct 55 SRKLAGLVLKNALDAKEQHRKYELVQRWLALDMSTKSQIRAFLLKTLSAPVPDVRSTASQ 114
Query 65 ALAKLGVIELRSLEFEDLLPALQDMVQQ 92
+AK+ IEL ++ +L+ +L + Q
Sbjct 115 VIAKVAGIELPQKQWPELIVSLLSNIHQ 142
> mmu:16211 Kpnb1, AA409963, IPOB, Impnb, MGC8315; karyopherin
(importin) beta 1; K14293 importin subunit beta-1
Length=876
Score = 37.0 bits (84), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 24/83 (28%), Positives = 44/83 (53%), Gaps = 1/83 (1%)
Query 4 IAKQIAAVTFKNSISAKDMTLDGAAAQAWRALPAPTKAEIKKQLLAALQTECTQVGNAVC 63
+A+ A + KNS+++KD + Q W A+ A + E+K +L L TE + +A
Sbjct 52 VARVAAGLQIKNSLTSKDPDIKAQYQQRWLAIDANARREVKNYVLQTLGTETYRPSSA-S 110
Query 64 QALAKLGVIELRSLEFEDLLPAL 86
Q +A + E+ ++ +L+P L
Sbjct 111 QCVAGIACAEIPVSQWPELIPQL 133
> hsa:3837 KPNB1, IMB1, IPO1, IPOB, Impnb, MGC2155, MGC2156, MGC2157,
NTF97; karyopherin (importin) beta 1; K14293 importin
subunit beta-1
Length=876
Score = 37.0 bits (84), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 24/83 (28%), Positives = 44/83 (53%), Gaps = 1/83 (1%)
Query 4 IAKQIAAVTFKNSISAKDMTLDGAAAQAWRALPAPTKAEIKKQLLAALQTECTQVGNAVC 63
+A+ A + KNS+++KD + Q W A+ A + E+K +L L TE + +A
Sbjct 52 VARVAAGLQIKNSLTSKDPDIKAQYQQRWLAIDANARREVKNYVLQTLGTETYRPSSA-S 110
Query 64 QALAKLGVIELRSLEFEDLLPAL 86
Q +A + E+ ++ +L+P L
Sbjct 111 QCVAGIACAEIPVNQWPELIPQL 133
> dre:570700 kpnb1, MGC123184, wu:fc12b10, zgc:123184; karyopherin
(importin) beta 1; K14293 importin subunit beta-1
Length=876
Score = 35.8 bits (81), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 24/83 (28%), Positives = 43/83 (51%), Gaps = 1/83 (1%)
Query 4 IAKQIAAVTFKNSISAKDMTLDGAAAQAWRALPAPTKAEIKKQLLAALQTECTQVGNAVC 63
+A+ A + KNS+++KD + Q W A+ + EIK +L L TE + +A
Sbjct 52 VARVAAGLQVKNSLTSKDPEVKSQYQQRWLAIDTSARREIKNYVLQTLGTETYRPSSA-S 110
Query 64 QALAKLGVIELRSLEFEDLLPAL 86
Q +A + E+ ++ +L+P L
Sbjct 111 QCVAGIACAEIPVNQWPELIPQL 133
> xla:100137718 kpnb1, imb1, impnb, ipo1, ipob, ntf97; karyopherin
(importin) beta 1; K14293 importin subunit beta-1
Length=876
Score = 35.0 bits (79), Expect = 0.066, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 42/83 (50%), Gaps = 1/83 (1%)
Query 4 IAKQIAAVTFKNSISAKDMTLDGAAAQAWRALPAPTKAEIKKQLLAALQTECTQVGNAVC 63
+A+ A + KN ++++D + Q W A+ A + EIK +L L TE + +A
Sbjct 52 VARVAAGLQIKNPLTSRDPDVKAQYQQRWLAIDASARGEIKTYVLRTLGTESYRPSSA-S 110
Query 64 QALAKLGVIELRSLEFEDLLPAL 86
Q +A + E+ ++ L+P L
Sbjct 111 QCVAGIACAEITVNQWPQLIPQL 133
> dre:192295 ccna2, CycA2, cb671, chunp6924, wu:fi36f03; cyclin
A2; K06627 cyclin A
Length=428
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 26/44 (59%), Gaps = 2/44 (4%)
Query 71 VIELRSLEFEDLLPALQDMVQQAVSCQQPQKQQQQKQQKQRGSK 114
+++L EDLLP +QD+ Q ++ Q QQ ++K +GSK
Sbjct 371 LVDLTGYSLEDLLPCVQDLHQTYLAAS--QHAQQAVREKYKGSK 412
> sce:YER110C KAP123, YRB4; Kap123p
Length=1113
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 30/59 (50%), Gaps = 1/59 (1%)
Query 29 AQAWRALPAPTKAEIKKQLL-AALQTECTQVGNAVCQALAKLGVIELRSLEFEDLLPAL 86
++ W A+ T+A IK LL A V ++ + +A +G EL ++ DL+P L
Sbjct 68 SKHWNAIDESTRASIKTSLLQTAFSEPKENVRHSNARVIASIGTEELDGNKWPDLVPNL 126
Lambda K H
0.318 0.127 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2044474180
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40