bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_3168_orf2
Length=146
Score E
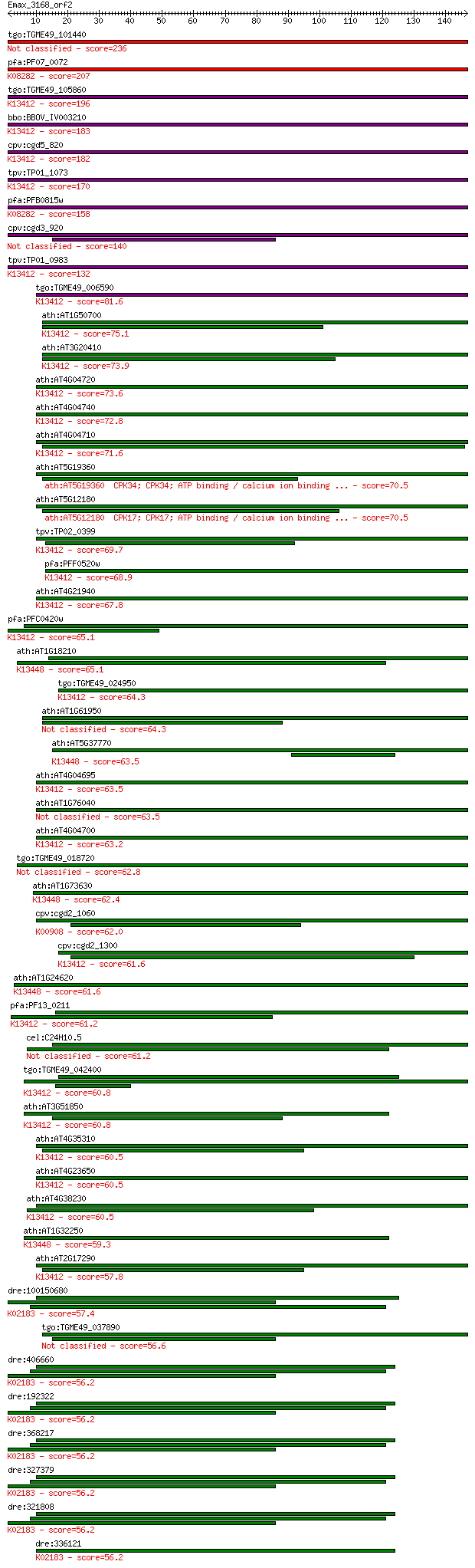
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_101440 calmodulin-domain protein kinase 1, putative... 236 2e-62
pfa:PF07_0072 PfCDPK4; calcium-dependent protein kinase 4 (EC:... 207 7e-54
tgo:TGME49_105860 calcium-dependent protein kinase 1, putative... 196 2e-50
bbo:BBOV_IV003210 21.m02835; calcium-dependent protein kinase ... 183 2e-46
cpv:cgd5_820 calcium/calmodulin dependent protein kinase with ... 182 3e-46
tpv:TP01_1073 calmodulin-domain protein kinase; K13412 calcium... 170 1e-42
pfa:PFB0815w CDPK1, CPK; Calcium-dependent protein kinase 1 (E... 158 7e-39
cpv:cgd3_920 calmodulin-domain protein kinase 1 140 1e-33
tpv:TP01_0983 calcium-dependent protein kinase; K13412 calcium... 132 4e-31
tgo:TGME49_006590 calcium-dependent protein kinase, putative (... 81.6 8e-16
ath:AT1G50700 CPK33; CPK33; ATP binding / calcium ion binding ... 75.1 7e-14
ath:AT3G20410 CPK9; CPK9 (calmodulin-domain protein kinase 9);... 73.9 2e-13
ath:AT4G04720 CPK21; CPK21; ATP binding / calcium ion binding ... 73.6 2e-13
ath:AT4G04740 CPK23; CPK23; ATP binding / calcium ion binding ... 72.8 3e-13
ath:AT4G04710 CPK22; CPK22; ATP binding / calcium ion binding ... 71.6 8e-13
ath:AT5G19360 CPK34; CPK34; ATP binding / calcium ion binding ... 70.5 2e-12
ath:AT5G12180 CPK17; CPK17; ATP binding / calcium ion binding ... 70.5 2e-12
tpv:TP02_0399 calcium-dependent protein kinase; K13412 calcium... 69.7 3e-12
pfa:PFF0520w PfCDPK2; calcium-dependent protein kinase (EC:2.7... 68.9 5e-12
ath:AT4G21940 CPK15; CPK15; ATP binding / calcium ion binding ... 67.8 1e-11
pfa:PFC0420w PfCDPK3; calcium-dependent protein kinase-3 (EC:2... 65.1 8e-11
ath:AT1G18210 calcium-binding protein, putative; K13448 calciu... 65.1 8e-11
tgo:TGME49_024950 protein kinase 6, putative (EC:1.6.3.1 2.7.1... 64.3 1e-10
ath:AT1G61950 CPK19; CPK19; ATP binding / calcium ion binding ... 64.3 1e-10
ath:AT5G37770 TCH2; TCH2 (TOUCH 2); calcium ion binding; K1344... 63.5 2e-10
ath:AT4G04695 CPK31; CPK31; ATP binding / calcium ion binding ... 63.5 2e-10
ath:AT1G76040 CPK29; CPK29; ATP binding / calcium ion binding ... 63.5 2e-10
ath:AT4G04700 CPK27; CPK27; ATP binding / calcium ion binding ... 63.2 3e-10
tgo:TGME49_018720 calcium-dependent protein kinase, putative (... 62.8 3e-10
ath:AT1G73630 calcium-binding protein, putative; K13448 calciu... 62.4 5e-10
cpv:cgd2_1060 calcium/calmodulin dependent protein kinase with... 62.0 6e-10
cpv:cgd2_1300 calcium/calmodulin dependent protein kinase with... 61.6 7e-10
ath:AT1G24620 polcalcin, putative / calcium-binding pollen all... 61.6 8e-10
pfa:PF13_0211 calcium-dependent protein kinase, putative (EC:2... 61.2 1e-09
cel:C24H10.5 uvt-2; Unidentified Vitellogenin-linked Transcrip... 61.2 1e-09
tgo:TGME49_042400 calcium-dependent protein kinase, putative (... 60.8 1e-09
ath:AT3G51850 CPK13; CPK13; ATP binding / calcium ion binding ... 60.8 2e-09
ath:AT4G35310 CPK5; CPK5 (calmodulin-domain protein kinase 5);... 60.5 2e-09
ath:AT4G23650 CDPK6; CDPK6 (CALCIUM-DEPENDENT PROTEIN KINASE 6... 60.5 2e-09
ath:AT4G38230 CPK26; CPK26; ATP binding / calcium ion binding ... 60.5 2e-09
ath:AT1G32250 calmodulin, putative; K13448 calcium-binding pro... 59.3 4e-09
ath:AT2G17290 CPK6; CPK6 (CALCIUM-DEPENDENT PROTEIN KINASE 6);... 57.8 1e-08
dre:100150680 calmodulin 2-like; K02183 calmodulin 57.4 2e-08
tgo:TGME49_037890 calcium-dependent protein kinase, putative (... 56.6 2e-08
dre:406660 calm1a, CaMbeta, sb:cb617, wu:fb08d09, wu:fb69c08, ... 56.2 3e-08
dre:192322 calm2b, calm2, cb169, zgc:64036; calmodulin 2b, (ph... 56.2 3e-08
dre:368217 calm1b, CaMbeta-2, zgc:73137; calmodulin 1b; K02183... 56.2 3e-08
dre:327379 calm3a, CaMgamma, MGC192068, calm2g, wu:fi05b08, zg... 56.2 3e-08
dre:321808 calm3b, wu:fb36a09, zgc:55591, zgc:76987; calmoduli... 56.2 3e-08
dre:336121 calm2a, CaMalpha, calm2d, cb815, wu:fj49d04, zgc:73... 56.2 3e-08
> tgo:TGME49_101440 calmodulin-domain protein kinase 1, putative
(EC:2.7.11.17)
Length=582
Score = 236 bits (601), Expect = 2e-62, Method: Compositional matrix adjust.
Identities = 111/146 (76%), Positives = 132/146 (90%), Gaps = 0/146 (0%)
Query 1 LLYMGSKLTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAI 60
LLYMGSKLT+ +ET EL IF KMDKNGDGQLD+ EL+EGY ELM++KG+D S+LD SA+
Sbjct 419 LLYMGSKLTSQDETKELTAIFHKMDKNGDGQLDRAELIEGYKELMRMKGQDASMLDASAV 478
Query 61 ETEVEQVLEAVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSEL 120
E EV+QVL+AVDFDKNG+IEYSEFVTVAMDR+TLLSR+RLERAF MFD+D SGKISS+EL
Sbjct 479 EHEVDQVLDAVDFDKNGYIEYSEFVTVAMDRKTLLSRERLERAFRMFDSDNSGKISSTEL 538
Query 121 ATIFGVSELDSEAWRRVLAEVDRNND 146
ATIFGVS++DSE W+ VL+EVD+NND
Sbjct 539 ATIFGVSDVDSETWKSVLSEVDKNND 564
> pfa:PF07_0072 PfCDPK4; calcium-dependent protein kinase 4 (EC:2.7.1.11);
K08282 non-specific serine/threonine protein kinase
[EC:2.7.11.1]
Length=528
Score = 207 bits (528), Expect = 7e-54, Method: Compositional matrix adjust.
Identities = 98/146 (67%), Positives = 126/146 (86%), Gaps = 0/146 (0%)
Query 1 LLYMGSKLTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAI 60
LLYMGSKLTT +ET EL KIF+KMDKNGDGQLD+ EL+ GY EL+KLKGED S LD +AI
Sbjct 365 LLYMGSKLTTIDETKELTKIFKKMDKNGDGQLDRNELIIGYKELLKLKGEDTSDLDNAAI 424
Query 61 ETEVEQVLEAVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSEL 120
E EV+Q+L ++D D+NG+IEYSEF+TV++DR+ LLS +RLE+AF++FD DGSGKIS++EL
Sbjct 425 EYEVDQILNSIDLDQNGYIEYSEFLTVSIDRKLLLSTERLEKAFKLFDKDGSGKISANEL 484
Query 121 ATIFGVSELDSEAWRRVLAEVDRNND 146
A +FG+S++ SE W+ VL EVD+NND
Sbjct 485 AQLFGLSDVSSECWKTVLKEVDQNND 510
> tgo:TGME49_105860 calcium-dependent protein kinase 1, putative
(EC:2.7.11.17); K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=537
Score = 196 bits (499), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 91/146 (62%), Positives = 121/146 (82%), Gaps = 0/146 (0%)
Query 1 LLYMGSKLTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAI 60
+L+MGSKLTT EET EL +IF+++D NGDGQLD++EL+EGY +LM+ KG+ VS LD S I
Sbjct 370 MLFMGSKLTTLEETKELTQIFRQLDNNGDGQLDRKELIEGYRKLMQWKGDTVSDLDSSQI 429
Query 61 ETEVEQVLEAVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSEL 120
E EV+ +L++VDFD+NG+IEYSEFVTV MD++ LLSR+RL AF+ FD+DGSGKI++ EL
Sbjct 430 EAEVDHILQSVDFDRNGYIEYSEFVTVCMDKQLLLSRERLLAAFQQFDSDGSGKITNEEL 489
Query 121 ATIFGVSELDSEAWRRVLAEVDRNND 146
+FGV+E+D E W +VL E D+NND
Sbjct 490 GRLFGVTEVDDETWHQVLQECDKNND 515
> bbo:BBOV_IV003210 21.m02835; calcium-dependent protein kinase
4 (EC:2.7.11.1); K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=517
Score = 183 bits (464), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 83/146 (56%), Positives = 116/146 (79%), Gaps = 0/146 (0%)
Query 1 LLYMGSKLTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAI 60
LLY+GSKL T EE++ L IF KMDKNGDGQLD+ EL++G+ E ++LKG ++ ++
Sbjct 356 LLYIGSKLVTKEESNYLTTIFSKMDKNGDGQLDRSELIDGFSEYLRLKGTAADNAERMSV 415
Query 61 ETEVEQVLEAVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSEL 120
E +V+Q+L+ +DFDKNG+I+YSEF+TVAMDRR L+ + RLE+AF++FD DGSG ISS EL
Sbjct 416 EEQVDQILQDIDFDKNGYIDYSEFLTVAMDRRNLMQKDRLEKAFKLFDADGSGTISSEEL 475
Query 121 ATIFGVSELDSEAWRRVLAEVDRNND 146
+FGV+++ ++ W+RVL EVD NND
Sbjct 476 GKMFGVADVSADDWQRVLHEVDSNND 501
> cpv:cgd5_820 calcium/calmodulin dependent protein kinase with
a kinase domain and 4 calmodulin like EF hands ; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=523
Score = 182 bits (462), Expect = 3e-46, Method: Compositional matrix adjust.
Identities = 86/146 (58%), Positives = 116/146 (79%), Gaps = 3/146 (2%)
Query 1 LLYMGSKLTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAI 60
LLYM SKLT+ EET EL IF+ +DKNGDGQLD+QEL++GY KL GE+V+V D I
Sbjct 355 LLYMASKLTSQEETKELTDIFRHIDKNGDGQLDRQELIDGY---SKLSGEEVAVFDLPQI 411
Query 61 ETEVEQVLEAVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSEL 120
E+EV+ +L A DFD+NG+I+YSEFVTVAMDR++LLS+ +LE AF+ FD DG+GKIS EL
Sbjct 412 ESEVDAILGAADFDRNGYIDYSEFVTVAMDRKSLLSKDKLESAFQKFDQDGNGKISVDEL 471
Query 121 ATIFGVSELDSEAWRRVLAEVDRNND 146
A++FG+ L+S+ W+ +++ +D NND
Sbjct 472 ASVFGLDHLESKTWKEMISGIDSNND 497
> tpv:TP01_1073 calmodulin-domain protein kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=504
Score = 170 bits (431), Expect = 1e-42, Method: Compositional matrix adjust.
Identities = 76/146 (52%), Positives = 108/146 (73%), Gaps = 0/146 (0%)
Query 1 LLYMGSKLTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAI 60
+LY+GSKL T +E L IF MDKNGDGQLD+ EL+EGY + +K KG+ + +++++ I
Sbjct 342 MLYIGSKLLTRDECKSLTAIFNSMDKNGDGQLDRSELIEGYTQYLKFKGKGMELMERTQI 401
Query 61 ETEVEQVLEAVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSEL 120
E +V+ +L+ +DFD NG+I+YSEF+TVAMDRRTL S++RLE AF +FD D SG I +EL
Sbjct 402 EEQVDTILQDIDFDNNGYIDYSEFLTVAMDRRTLFSKERLENAFRLFDFDNSGTICCNEL 461
Query 121 ATIFGVSELDSEAWRRVLAEVDRNND 146
A +FG + E+W+ ++ E D NND
Sbjct 462 AKMFGADDGGKESWQHLIGEFDTNND 487
> pfa:PFB0815w CDPK1, CPK; Calcium-dependent protein kinase 1
(EC:2.7.11.1); K08282 non-specific serine/threonine protein
kinase [EC:2.7.11.1]
Length=524
Score = 158 bits (399), Expect = 7e-39, Method: Compositional matrix adjust.
Identities = 78/146 (53%), Positives = 105/146 (71%), Gaps = 3/146 (2%)
Query 1 LLYMGSKLTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAI 60
+L++GSKLTT EE EL IF+K+DKNGDGQLDK+EL+EGY L K E + + +
Sbjct 361 ILFIGSKLTTLEERKELTDIFKKLDKNGDGQLDKKELIEGYNILRSFKNE---LGELKNV 417
Query 61 ETEVEQVLEAVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSEL 120
E EV+ +L+ VDFDKNG+IEYSEF++V MD++ L S +RL AF +FDTD SGKI+ EL
Sbjct 418 EEEVDNILKEVDFDKNGYIEYSEFISVCMDKQILFSEERLRDAFNLFDTDKSGKITKEEL 477
Query 121 ATIFGVSELDSEAWRRVLAEVDRNND 146
A +FG++ + + W VL E D+N D
Sbjct 478 ANLFGLTSISEQMWNEVLGEADKNKD 503
> cpv:cgd3_920 calmodulin-domain protein kinase 1
Length=538
Score = 140 bits (354), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 75/151 (49%), Positives = 103/151 (68%), Gaps = 5/151 (3%)
Query 1 LLYMGSKLTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVL---DK 57
LLYM SKLTT +ET +L +IF+K+D N DG LD+ EL+ GY E M+LKG D + L +
Sbjct 369 LLYMASKLTTLDETKQLTEIFRKLDTNNDGMLDRDELVRGYHEFMRLKGVDSNSLIQNEG 428
Query 58 SAIETEVEQVLEAVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISS 117
S IE +++ ++ +D D +G IEYSEF+ A+DR LLSR+R+ERAF+MFD DGSGKIS+
Sbjct 429 STIEDQIDSLMPLLDMDGSGSIEYSEFIASAIDRTILLSRERMERAFKMFDKDGSGKIST 488
Query 118 SELATIFGV--SELDSEAWRRVLAEVDRNND 146
EL +F S + E ++ +VD N D
Sbjct 489 KELFKLFSQADSSIQMEELESIIEQVDNNKD 519
Score = 33.5 bits (75), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 20/71 (28%), Positives = 39/71 (54%), Gaps = 11/71 (15%)
Query 15 DELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLEAVDFD 74
+ + + F+ DK+G G++ +EL + + S D S E+E ++E VD +
Sbjct 469 ERMERAFKMFDKDGSGKISTKELFKLF-----------SQADSSIQMEELESIIEQVDNN 517
Query 75 KNGFIEYSEFV 85
K+G ++++EFV
Sbjct 518 KDGEVDFNEFV 528
> tpv:TP01_0983 calcium-dependent protein kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=509
Score = 132 bits (332), Expect = 4e-31, Method: Composition-based stats.
Identities = 66/146 (45%), Positives = 102/146 (69%), Gaps = 1/146 (0%)
Query 1 LLYMGSKLTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAI 60
+L M +K TTN+E +EL+K+F ++D NGDG LD+ EL++GY + + + S + I
Sbjct 347 MLLMANKFTTNDEINELSKLFSQLDTNGDGALDRSELIQGYKSIKQNLRDGCSRMSNEEI 406
Query 61 ETEVEQVLEAVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSEL 120
E EV++++ + D D +G I+YSEF+T +D+ TLLS++RL+ AF FD DGSGKIS+SEL
Sbjct 407 EKEVDEIIRSCDLDHSGSIDYSEFITGCIDKLTLLSKERLKLAFSTFDVDGSGKISNSEL 466
Query 121 ATIFGVSELDSEAWRRVLAEVDRNND 146
+ I GVS + + ++L E+D N+D
Sbjct 467 SQILGVSNAPN-LFNQILNEIDTNHD 491
> tgo:TGME49_006590 calcium-dependent protein kinase, putative
(EC:2.7.11.17 3.4.22.53); K13412 calcium-dependent protein
kinase [EC:2.7.11.1]
Length=761
Score = 81.6 bits (200), Expect = 8e-16, Method: Composition-based stats.
Identities = 46/141 (32%), Positives = 75/141 (53%), Gaps = 15/141 (10%)
Query 10 TNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLE 69
+ +E D L +IF +D + G L QE+ EG + L + I +++ ++E
Sbjct 521 SEKEIDHLRQIFMTLDVDNSGTLSVQEVREG-----------LKRLGWTEIPADLQAIIE 569
Query 70 AVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATIFGVSEL 129
VD DK+G I+Y+EF+ MD++ + AF +FD DG+GKIS EL + G+ ++
Sbjct 570 EVDSDKSGHIDYTEFIAATMDKKLYMKEDVCWAAFRVFDLDGNGKISQDELKKVLGMPDV 629
Query 130 DSEAWR----RVLAEVDRNND 146
++ R +L EVD N D
Sbjct 630 ETAVGRATIDALLTEVDLNGD 650
> ath:AT1G50700 CPK33; CPK33; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=521
Score = 75.1 bits (183), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 48/138 (34%), Positives = 75/138 (54%), Gaps = 16/138 (11%)
Query 12 EETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLEAV 71
EE L +F +D + G + +EL EG +L G ++ E EV+Q+++A
Sbjct 374 EEIQGLKAMFANIDTDNSGTITYEELKEGLAKL----GSRLT-------EAEVKQLMDAA 422
Query 72 DFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATI---FGVSE 128
D D NG I+Y EF+T M R L S + + +AF+ FD DGSG I++ EL +G+
Sbjct 423 DVDGNGSIDYIEFITATMHRHRLESNENVYKAFQHFDKDGSGYITTDELEAALKEYGMG- 481
Query 129 LDSEAWRRVLAEVDRNND 146
D + +L++VD +ND
Sbjct 482 -DDATIKEILSDVDADND 498
Score = 31.2 bits (69), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 27/89 (30%), Positives = 39/89 (43%), Gaps = 14/89 (15%)
Query 12 EETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLEAV 71
E + + K FQ DK+G G + EL E G+D ++ +++L V
Sbjct 446 ESNENVYKAFQHFDKDGSGYITTDELEAALKEYGM--GDDATI----------KEILSDV 493
Query 72 DFDKNGFIEYSEFVTVAMDRRTLLSRQRL 100
D D +G I Y EF AM R + RL
Sbjct 494 DADNDGRINYDEF--CAMMRSGNPQQPRL 520
> ath:AT3G20410 CPK9; CPK9 (calmodulin-domain protein kinase 9);
calmodulin-dependent protein kinase/ kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=541
Score = 73.9 bits (180), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 49/138 (35%), Positives = 74/138 (53%), Gaps = 16/138 (11%)
Query 12 EETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLEAV 71
EE L +F +D + G + +EL EG +L G ++ E EV+Q+++A
Sbjct 392 EEIQGLKAMFANIDTDNSGTITYEELKEGLAKL----GSKLT-------EAEVKQLMDAA 440
Query 72 DFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATI---FGVSE 128
D D NG I+Y EF+T M R L S + L +AF+ FD D SG I+ EL + +G+
Sbjct 441 DVDGNGSIDYIEFITATMHRHRLESNENLYKAFQHFDKDSSGYITIDELESALKEYGMG- 499
Query 129 LDSEAWRRVLAEVDRNND 146
D + VL++VD +ND
Sbjct 500 -DDATIKEVLSDVDSDND 516
Score = 29.3 bits (64), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 27/93 (29%), Positives = 40/93 (43%), Gaps = 15/93 (16%)
Query 12 EETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLEAV 71
E + L K FQ DK+ G + EL E G+D ++ ++VL V
Sbjct 464 ESNENLYKAFQHFDKDSSGYITIDELESALKEYGM--GDDATI----------KEVLSDV 511
Query 72 DFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAF 104
D D +G I Y EF + R+ +Q+ R F
Sbjct 512 DSDNDGRINYEEFCAMM---RSGNPQQQQPRLF 541
> ath:AT4G04720 CPK21; CPK21; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=531
Score = 73.6 bits (179), Expect = 2e-13, Method: Composition-based stats.
Identities = 48/138 (34%), Positives = 72/138 (52%), Gaps = 12/138 (8%)
Query 10 TNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLE 69
+ EE L +F +D + G + +EL G L G +S ETEV+Q++E
Sbjct 378 SEEEIKGLKTMFANIDTDKSGTITYEELKTGLTRL----GSRLS-------ETEVKQLME 426
Query 70 AVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATIFGVSEL 129
A D D NG I+Y EF++ M R L + + +AF+ FD D SG I+ EL + +
Sbjct 427 AADVDGNGTIDYYEFISATMHRYKLDRDEHVYKAFQHFDKDNSGHITRDELESAMKEYGM 486
Query 130 DSEA-WRRVLAEVDRNND 146
EA + V++EVD +ND
Sbjct 487 GDEASIKEVISEVDTDND 504
> ath:AT4G04740 CPK23; CPK23; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=520
Score = 72.8 bits (177), Expect = 3e-13, Method: Composition-based stats.
Identities = 46/138 (33%), Positives = 70/138 (50%), Gaps = 12/138 (8%)
Query 10 TNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLE 69
+ EE L +F MD N G + ++L G +S L ETEV+Q++E
Sbjct 367 SEEEIKGLKTLFANMDTNRSGTITYEQLQTG-----------LSRLRSRLSETEVQQLVE 415
Query 70 AVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATIFGVSEL 129
A D D NG I+Y EF++ M R L + + +AF+ D D +G I+ EL + +
Sbjct 416 ASDVDGNGTIDYYEFISATMHRYKLHHDEHVHKAFQHLDKDKNGHITRDELESAMKEYGM 475
Query 130 DSEA-WRRVLAEVDRNND 146
EA + V++EVD +ND
Sbjct 476 GDEASIKEVISEVDTDND 493
> ath:AT4G04710 CPK22; CPK22; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=575
Score = 71.6 bits (174), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 49/140 (35%), Positives = 74/140 (52%), Gaps = 14/140 (10%)
Query 10 TNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLE 69
+ EE L +F+ MD + G + +EL G + G +S ETEV+Q++E
Sbjct 330 SEEEIKGLKTMFENMDMDKSGSITYEELKMG----LNRHGSKLS-------ETEVKQLME 378
Query 70 AV--DFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATIFGVS 127
AV D D NG I+Y EF++ M R L + L +AF+ FD DGSG I+ E+
Sbjct 379 AVSADVDGNGTIDYIEFISATMHRHRLERDEHLYKAFQYFDKDGSGHITKEEVEIAMKEH 438
Query 128 ELDSEA-WRRVLAEVDRNND 146
+ EA + +++E D+NND
Sbjct 439 GMGDEANAKDLISEFDKNND 458
Score = 41.6 bits (96), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 45/166 (27%), Positives = 70/166 (42%), Gaps = 49/166 (29%)
Query 12 EETDELNKIFQKMDKNGDGQLDKQELMEGYVEL-MKLKGEDVSVLDKSAIETEVEQVLEA 70
E + L K FQ DK+G G + K+E VE+ MK G E + ++
Sbjct 406 ERDEHLYKAFQYFDKDGSGHITKEE-----VEIAMKEHG--------MGDEANAKDLISE 452
Query 71 VDFDKNGFIEYSEFVTVAMDRRTLLSRQ----------------------------RLER 102
D + +G I+Y EF T M R +L Q +++
Sbjct 453 FDKNNDGKIDYEEFCT--MMRNGILQPQGKLLKRLYMNLEELKTGLTRLGSRLSETEIDK 510
Query 103 AFEMFDTDGSGKISSSELATI---FGVSELDSEAWRRVLAEVDRNN 145
AF+ FD D SG I+ EL + +G+ D + + V++EVD +N
Sbjct 511 AFQHFDKDNSGHITRDELESAMKEYGMG--DEASIKEVISEVDTDN 554
> ath:AT5G19360 CPK34; CPK34; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase; K00924 [EC:2.7.1.-]
Length=523
Score = 70.5 bits (171), Expect = 2e-12, Method: Composition-based stats.
Identities = 47/140 (33%), Positives = 74/140 (52%), Gaps = 16/140 (11%)
Query 10 TNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLE 69
+ EE L ++F+ MD + G + +EL +G + +G +S E EV+Q++E
Sbjct 367 SEEEIMGLKEMFKGMDTDNSGTITLEELRQGLAK----QGTRLS-------EYEVQQLME 415
Query 70 AVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATI---FGV 126
A D D NG I+Y EF+ M L + L AF+ FD D SG I++ EL FG+
Sbjct 416 AADADGNGTIDYGEFIAATMHINRLDREEHLYSAFQHFDKDNSGYITTEELEQALREFGM 475
Query 127 SELDSEAWRRVLAEVDRNND 146
+ D + +++EVD +ND
Sbjct 476 N--DGRDIKEIISEVDGDND 493
Score = 33.1 bits (74), Expect = 0.28, Method: Composition-based stats.
Identities = 23/81 (28%), Positives = 36/81 (44%), Gaps = 14/81 (17%)
Query 12 EETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLEAV 71
+ + L FQ DK+ G + +EL + E G D+ ++++ V
Sbjct 441 DREEHLYSAFQHFDKDNSGYITTEELEQALREFGMNDGRDI------------KEIISEV 488
Query 72 DFDKNGFIEYSEFVTVAMDRR 92
D D +G I Y EF VAM R+
Sbjct 489 DGDNDGRINYEEF--VAMMRK 507
> ath:AT5G12180 CPK17; CPK17; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase; K00924 [EC:2.7.1.-]
Length=528
Score = 70.5 bits (171), Expect = 2e-12, Method: Composition-based stats.
Identities = 47/140 (33%), Positives = 73/140 (52%), Gaps = 16/140 (11%)
Query 10 TNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLE 69
+ EE L ++F+ MD + G + +EL +G + +G +S E EV+Q++E
Sbjct 372 SEEEIMGLKEMFKGMDTDSSGTITLEELRQGLAK----QGTRLS-------EYEVQQLME 420
Query 70 AVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATI---FGV 126
A D D NG I+Y EF+ M L + L AF+ FD D SG I+ EL FG+
Sbjct 421 AADADGNGTIDYGEFIAATMHINRLDREEHLYSAFQHFDKDNSGYITMEELEQALREFGM 480
Query 127 SELDSEAWRRVLAEVDRNND 146
+ D + +++EVD +ND
Sbjct 481 N--DGRDIKEIISEVDGDND 498
Score = 33.5 bits (75), Expect = 0.21, Method: Composition-based stats.
Identities = 26/97 (26%), Positives = 44/97 (45%), Gaps = 17/97 (17%)
Query 12 EETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLEAV 71
+ + L FQ DK+ G + +EL + E G D+ ++++ V
Sbjct 446 DREEHLYSAFQHFDKDNSGYITMEELEQALREFGMNDGRDI------------KEIISEV 493
Query 72 DFDKNGFIEYSEFVTVAMDRR---TLLSRQRLERAFE 105
D D +G I Y EF VAM R+ + ++R E +F+
Sbjct 494 DGDNDGRINYDEF--VAMMRKGNPDPIPKKRRELSFK 528
> tpv:TP02_0399 calcium-dependent protein kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=844
Score = 69.7 bits (169), Expect = 3e-12, Method: Composition-based stats.
Identities = 44/141 (31%), Positives = 69/141 (48%), Gaps = 17/141 (12%)
Query 10 TNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLE 69
+ E L F+ +DK+GDG L E+ G LK S +EQ+++
Sbjct 698 SERELAPLTNAFESLDKDGDGVLSLDEVANG------LKHSKQSSF-------HIEQIVK 744
Query 70 AVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATIFGVS-- 127
+D D++G IEY+EFV A+D R + +RAF +FDTD G+I+ ++ +F
Sbjct 745 GIDTDQSGIIEYTEFVAAAIDARLYNQKDFFKRAFNIFDTDRDGRITREDMYRVFSTEST 804
Query 128 --ELDSEAWRRVLAEVDRNND 146
+ E +L EVD + D
Sbjct 805 NPRMTQEMVEDILEEVDLDRD 825
Score = 33.5 bits (75), Expect = 0.25, Method: Composition-based stats.
Identities = 18/79 (22%), Positives = 36/79 (45%), Gaps = 9/79 (11%)
Query 13 ETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLEAVD 72
+ D + F D + DG++ ++++ + + + VE +LE VD
Sbjct 771 QKDFFKRAFNIFDTDRDGRITREDMYRVF---------STESTNPRMTQEMVEDILEEVD 821
Query 73 FDKNGFIEYSEFVTVAMDR 91
D++G I Y EF ++ +R
Sbjct 822 LDRDGTISYDEFTSMLSNR 840
> pfa:PFF0520w PfCDPK2; calcium-dependent protein kinase (EC:2.7.11.1);
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=509
Score = 68.9 bits (167), Expect = 5e-12, Method: Composition-based stats.
Identities = 44/138 (31%), Positives = 73/138 (52%), Gaps = 15/138 (10%)
Query 13 ETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLEAVD 72
E + L IF +D + G L QE+++G ++ K I ++ QVL +D
Sbjct 371 EINNLRNIFIALDVDNSGTLSSQEILDGLKKIGYQK-----------IPPDIHQVLRDID 419
Query 73 FDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATIFGVSELDS- 131
+ +G I Y++F+ +D++T L ++ F+ FD DG+GKIS EL IFG ++++
Sbjct 420 SNASGQIHYTDFLAATIDKQTYLKKEVCLIPFKFFDIDGNGKISVEELKRIFGRDDIENP 479
Query 132 ---EAWRRVLAEVDRNND 146
+A +L EVD N D
Sbjct 480 LIDKAIDSLLQEVDLNGD 497
> ath:AT4G21940 CPK15; CPK15; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=554
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 47/140 (33%), Positives = 73/140 (52%), Gaps = 16/140 (11%)
Query 10 TNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLE 69
+ EE L +F MD + G + +EL G +L G ++ E EV+Q++E
Sbjct 400 SEEEIKGLKTMFANMDTDKSGTITYEELKNGLAKL----GSKLT-------EAEVKQLME 448
Query 70 AVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATI---FGV 126
A D D NG I+Y EF++ M R + + +AF+ FD D SG I+ EL + +G+
Sbjct 449 AADVDGNGTIDYIEFISATMHRYRFDRDEHVFKAFQYFDKDNSGFITMDELESAMKEYGM 508
Query 127 SELDSEAWRRVLAEVDRNND 146
D + + V+AEVD +ND
Sbjct 509 G--DEASIKEVIAEVDTDND 526
> pfa:PFC0420w PfCDPK3; calcium-dependent protein kinase-3 (EC:2.7.11.17);
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=562
Score = 65.1 bits (157), Expect = 8e-11, Method: Composition-based stats.
Identities = 40/148 (27%), Positives = 82/148 (55%), Gaps = 20/148 (13%)
Query 6 SKLTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVE 65
++ + + + ++L F +D++G G + K++L +G + D + +
Sbjct 412 AQQSNDYDVEKLKSTFLVLDEDGKGYITKEQLKKGLEK------------DGLKLPYNFD 459
Query 66 QVLEAVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATIF- 124
+L+ +D D +G I+Y+EF+ A+DR+ LS++ + AF +FD D G+I+++ELA I
Sbjct 460 LLLDQIDSDGSGKIDYTEFIAAALDRKQ-LSKKLIYCAFRVFDVDNDGEITTAELAHILY 518
Query 125 ------GVSELDSEAWRRVLAEVDRNND 146
+++ D +R++ +VD+NND
Sbjct 519 NGNKKGNITQRDVNRVKRMIRDVDKNND 546
Score = 30.0 bits (66), Expect = 2.8, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 30/50 (60%), Gaps = 6/50 (12%)
Query 1 LLYMGSKL--TTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLK 48
+LY G+K T + + + ++ + +DKN DG++D E + E+MKLK
Sbjct 516 ILYNGNKKGNITQRDVNRVKRMIRDVDKNNDGKIDFHE----FSEMMKLK 561
> ath:AT1G18210 calcium-binding protein, putative; K13448 calcium-binding
protein CML
Length=170
Score = 65.1 bits (157), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 46/136 (33%), Positives = 70/136 (51%), Gaps = 18/136 (13%)
Query 14 TDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLEAVDF 73
+EL K+F + D NGDG++ EL G + S ETE+ +VLE VD
Sbjct 21 PEELKKVFDQFDSNGDGKISVLEL-----------GGVFKAMGTSYTETELNRVLEEVDT 69
Query 74 DKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATI---FGVSELD 130
D++G+I EF T+ R+ S + AF+++D D +G IS+SEL + G+S
Sbjct 70 DRDGYINLDEFSTLC---RSSSSAAEIRDAFDLYDQDKNGLISASELHQVLNRLGMS-CS 125
Query 131 SEAWRRVLAEVDRNND 146
E R++ VD + D
Sbjct 126 VEDCTRMIGPVDADGD 141
Score = 30.8 bits (68), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 30/117 (25%), Positives = 47/117 (40%), Gaps = 18/117 (15%)
Query 4 MGSKLTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETE 63
MG+ T ET ELN++ +++D + DG ++ E ++ S+ E
Sbjct 51 MGTSYT---ET-ELNRVLEEVDTDRDGYINLDEF--------------STLCRSSSSAAE 92
Query 64 VEQVLEAVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSEL 120
+ + D DKNG I SE V S + R D DG G ++ E
Sbjct 93 IRDAFDLYDQDKNGLISASELHQVLNRLGMSCSVEDCTRMIGPVDADGDGNVNFEEF 149
> tgo:TGME49_024950 protein kinase 6, putative (EC:1.6.3.1 2.7.11.17);
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=681
Score = 64.3 bits (155), Expect = 1e-10, Method: Composition-based stats.
Identities = 40/134 (29%), Positives = 63/134 (47%), Gaps = 17/134 (12%)
Query 17 LNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLEAVDFDKN 76
L+ F +D N DG L E+ +G + + GE E+ +L+ +D D N
Sbjct 522 LHDAFAALDTNADGVLTVAEIQQGLKQCC-VAGE------------EINDILKEMDTDGN 568
Query 77 GFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATIFGV----SELDSE 132
G I+Y+EF+ ++D + + AF++FD DG GKI+ EL + E
Sbjct 569 GTIDYTEFIAASIDHKIYEQESACQAAFKVFDLDGDGKITVDELQKVLETRCVQEAFSKE 628
Query 133 AWRRVLAEVDRNND 146
A ++ E D NND
Sbjct 629 AVAEMMKEGDSNND 642
> ath:AT1G61950 CPK19; CPK19; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase
Length=551
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 47/136 (34%), Positives = 66/136 (48%), Gaps = 12/136 (8%)
Query 12 EETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLEAV 71
EE L +F MD + G + EL G + L ETEV+Q+LE
Sbjct 400 EELKGLKTMFANMDTDKSGTITYDELKSG-----------LEKLGSRLTETEVKQLLEDA 448
Query 72 DFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATIFGVSEL-D 130
D D NG I+Y EF++ M+R + L +AF+ FD D SG IS EL T + D
Sbjct 449 DVDGNGTIDYIEFISATMNRFRVEREDNLFKAFQHFDKDNSGFISRQELETAMKEYNMGD 508
Query 131 SEAWRRVLAEVDRNND 146
+ +++EVD +ND
Sbjct 509 DIMIKEIISEVDADND 524
Score = 32.0 bits (71), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 35/76 (46%), Gaps = 12/76 (15%)
Query 12 EETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLEAV 71
E D L K FQ DK+ G + +QEL E G+D+ + ++++ V
Sbjct 472 EREDNLFKAFQHFDKDNSGFISRQELETAMKEYNM--GDDIMI----------KEIISEV 519
Query 72 DFDKNGFIEYSEFVTV 87
D D +G I Y EF +
Sbjct 520 DADNDGSINYQEFCNM 535
> ath:AT5G37770 TCH2; TCH2 (TOUCH 2); calcium ion binding; K13448
calcium-binding protein CML
Length=161
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 43/139 (30%), Positives = 73/139 (52%), Gaps = 18/139 (12%)
Query 15 DELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLEAVDFD 74
D++ K+FQ+ DKNGDG++ EL E + L +A E +++ D D
Sbjct 16 DDIKKVFQRFDKNGDGKISVDELKEV-----------IRALSPTASPEETVTMMKQFDLD 64
Query 75 KNGFIEYSEFVTVAMDRRTLLSRQR-----LERAFEMFDTDGSGKISSSELATIF-GVSE 128
NGFI+ EFV + R L+ AFE++D DG+G+IS+ EL ++ + E
Sbjct 65 GNGFIDLDEFVALFQIGIGGGGNNRNDVSDLKEAFELYDLDGNGRISAKELHSVMKNLGE 124
Query 129 LDS-EAWRRVLAEVDRNND 146
S + ++++++VD + D
Sbjct 125 KCSVQDCKKMISKVDIDGD 143
Score = 28.1 bits (61), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 91 RRTLLSRQRLERAFEMFDTDGSGKISSSELATI 123
R L S +++ F+ FD +G GKIS EL +
Sbjct 9 RSCLGSMDDIKKVFQRFDKNGDGKISVDELKEV 41
> ath:AT4G04695 CPK31; CPK31; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase/ protein tyrosine kinase;
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=484
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 46/140 (32%), Positives = 70/140 (50%), Gaps = 16/140 (11%)
Query 10 TNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLE 69
+ EE L +F +D + G + +EL G L G ++S +TEVEQ++E
Sbjct 330 SEEEIKGLKTLFTNIDTDKSGTITLEELKTGLTRL----GSNLS-------KTEVEQLME 378
Query 70 AVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATIF---GV 126
A D D NG I+ EF++ M R L + +AF+ FD D G I+ EL GV
Sbjct 379 AADVDGNGTIDIDEFISATMHRYRLDRDDHVYQAFQHFDKDNDGHITKEELEMAMKEHGV 438
Query 127 SELDSEAWRRVLAEVDRNND 146
D + ++++ EVD +ND
Sbjct 439 G--DEVSIKQIITEVDTDND 456
> ath:AT1G76040 CPK29; CPK29; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase/ protein tyrosine kinase
Length=323
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 48/140 (34%), Positives = 69/140 (49%), Gaps = 16/140 (11%)
Query 10 TNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLE 69
+ EE L + F+ MD + G + EL G + L E+E++Q++E
Sbjct 172 SEEEIKGLKQTFKNMDTDESGTITFDELRNG-----------LHRLGSKLTESEIKQLME 220
Query 70 AVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELA---TIFGV 126
A D DK+G I+Y EFVT M R L + L AF+ FD D SG I+ EL T +G+
Sbjct 221 AADVDKSGTIDYIEFVTATMHRHRLEKEENLIEAFKYFDKDRSGFITRDELKHSMTEYGM 280
Query 127 SELDSEAWRRVLAEVDRNND 146
D V+ +VD +ND
Sbjct 281 G--DDATIDEVINDVDTDND 298
> ath:AT4G04700 CPK27; CPK27; ATP binding / calcium ion binding
/ kinase/ protein kinase/ protein serine/threonine kinase/
protein tyrosine kinase; K13412 calcium-dependent protein kinase
[EC:2.7.11.1]
Length=485
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 44/140 (31%), Positives = 70/140 (50%), Gaps = 16/140 (11%)
Query 10 TNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLE 69
+ EE L +F +D + G + +EL G L G ++S +TEVEQ++E
Sbjct 330 SEEEIKGLKTLFTNIDTDKSGNITLEELKTGLTRL----GSNLS-------KTEVEQLME 378
Query 70 AVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATIF---GV 126
A D D NG I+ EF++ M R L + + +AF+ FD D G I+ EL G
Sbjct 379 AADMDGNGTIDIDEFISATMHRYKLDRDEHVYKAFQHFDKDNDGHITKEELEMAMKEDGA 438
Query 127 SELDSEAWRRVLAEVDRNND 146
D + ++++A+ D +ND
Sbjct 439 G--DEGSIKQIIADADTDND 456
> tgo:TGME49_018720 calcium-dependent protein kinase, putative
(EC:2.7.11.17)
Length=557
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 44/148 (29%), Positives = 76/148 (51%), Gaps = 20/148 (13%)
Query 4 MGSKL-TTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIET 62
M +L T ++ +N+IF+++DKNGDG L QEL EG E +
Sbjct 410 MAHQLNVTGQQIRHINQIFRQLDKNGDGLLSHQELTEGLAE-------------AGVPQW 456
Query 63 EVEQVLEAVDFDKNGFIEYSEFVTVAMD-RRTLLSRQRLERAFEMFDTDGSGKISSSELA 121
++ ++L+++D D +G + Y+EF+ + T L+ + AF+ D DG G+IS E
Sbjct 457 DINRILQSIDVDDSGNVSYTEFLAACYCWQETELN--VVWTAFQKIDKDGDGRISVREFC 514
Query 122 -TIFGVSE--LDSEAWRRVLAEVDRNND 146
+ G + E R ++A++DR+ D
Sbjct 515 DLVLGRDNKLIPEEELRAMVAQMDRDGD 542
> ath:AT1G73630 calcium-binding protein, putative; K13448 calcium-binding
protein CML
Length=163
Score = 62.4 bits (150), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 46/142 (32%), Positives = 71/142 (50%), Gaps = 19/142 (13%)
Query 9 TTNEETD-ELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQV 67
+T TD EL K+F K D NGDG++ EL G + S E E+ +V
Sbjct 12 STTPSTDMELKKVFDKFDANGDGKISVSEL-----------GNVFKSMGTSYTEEELNRV 60
Query 68 LEAVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATI---F 124
L+ +D D +GFI EF T+ R+ S + AF+++D + +G ISSSE+ +
Sbjct 61 LDEIDIDCDGFINQEEFATIC---RSSSSAVEIREAFDLYDQNKNGLISSSEIHKVLNRL 117
Query 125 GVSELDSEAWRRVLAEVDRNND 146
G++ E R++ VD + D
Sbjct 118 GMT-CSVEDCVRMIGHVDTDGD 138
> cpv:cgd2_1060 calcium/calmodulin dependent protein kinase with
a kinas domain and 4 calmodulin-like EF hands ; K00908 Ca2+/calmodulin-dependent
protein kinase [EC:2.7.11.17]
Length=718
Score = 62.0 bits (149), Expect = 6e-10, Method: Composition-based stats.
Identities = 41/146 (28%), Positives = 73/146 (50%), Gaps = 20/146 (13%)
Query 10 TNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLE 69
T + L + F +D N DG L QE++ G LK ++ L +++ +L
Sbjct 567 TESQISNLKEAFILLDANCDGTLTPQEIITG------LKNSGITEL-----PSDLLAILN 615
Query 70 AVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATIF----- 124
+D D +G I+Y+EF+ +D + Q AF++FD DG+GKI+++EL +F
Sbjct 616 DIDSDGSGSIDYTEFIAATLDSKQYSKEQVCWAAFKVFDQDGNGKITANELLNVFSYNSE 675
Query 125 ----GVSELDSEAWRRVLAEVDRNND 146
G+++ + ++ EVD + D
Sbjct 676 QGSAGINDKALSDVKNMIKEVDVDGD 701
Score = 29.3 bits (64), Expect = 4.0, Method: Composition-based stats.
Identities = 19/73 (26%), Positives = 38/73 (52%), Gaps = 6/73 (8%)
Query 21 FQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLEAVDFDKNGFIE 80
F+ D++G+G++ EL+ + E S ++V+ +++ VD D +G I+
Sbjct 650 FKVFDQDGNGKITANELLN----VFSYNSEQGSAGINDKALSDVKNMIKEVDVDGDGEID 705
Query 81 YSEFVTVAMDRRT 93
+ EF + M RR+
Sbjct 706 FQEF--LEMFRRS 716
> cpv:cgd2_1300 calcium/calmodulin dependent protein kinase with
a kinase domain and 4 calmodulin like EF hands ; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=677
Score = 61.6 bits (148), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 42/135 (31%), Positives = 72/135 (53%), Gaps = 16/135 (11%)
Query 17 LNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLEAVDFDKN 76
L K+F +D+NGDG L E+ L K++ + S + +++ +L +D D N
Sbjct 495 LQKLFSTLDRNGDGVLTINEIRSA---LHKIQ-------NVSQLGDDIDNLLMELDTDGN 544
Query 77 GFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATIFGVSELDSEAWRR 136
G I+Y+EF+ ++D + + AF++FD D G+IS EL+ + ++ L EA+ +
Sbjct 545 GRIDYTEFIAASIDHKLYEQESLCKAAFKVFDLDMDGRISPQELSRVLNITFL-QEAFEQ 603
Query 137 -----VLAEVDRNND 146
+L EVD N D
Sbjct 604 STIDSLLKEVDINQD 618
Score = 37.7 bits (86), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 26/112 (23%), Positives = 61/112 (54%), Gaps = 15/112 (13%)
Query 21 FQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLEAVDFDKNGFIE 80
F+ D + DG++ QEL + +++ L ++ ++ ++ +L+ VD +++G+I+
Sbjct 572 FKVFDLDMDGRISPQELS---------RVLNITFLQEAFEQSTIDSLLKEVDINQDGYID 622
Query 81 YSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSE---LATIFGVSEL 129
++EF+ + M + Q+LE + + +GSG S+ ++ IFG + +
Sbjct 623 FNEFMKMMMGDK---HEQKLETKVQKIEDEGSGNKKLSKGGIISDIFGATSI 671
> ath:AT1G24620 polcalcin, putative / calcium-binding pollen allergen,
putative; K13448 calcium-binding protein CML
Length=186
Score = 61.6 bits (148), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 44/149 (29%), Positives = 74/149 (49%), Gaps = 18/149 (12%)
Query 3 YMGSKLTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIET 62
Y+ + E EL +F+K D NGDG++ +EL G ++ L E
Sbjct 24 YLQKARSGKTEIRELEAVFKKFDVNGDGKISSKEL-----------GAIMTSLGHEVPEE 72
Query 63 EVEQVLEAVDFDKNGFIEYSEFV---TVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSE 119
E+E+ + +D +G+I + EFV T MD+ +L + L+ AF ++D DG+G IS+ E
Sbjct 73 ELEKAITEIDRKGDGYINFEEFVELNTKGMDQNDVL--ENLKDAFSVYDIDGNGSISAEE 130
Query 120 LATIFGV--SELDSEAWRRVLAEVDRNND 146
L + E R+++ VD++ D
Sbjct 131 LHEVLRSLGDECSIAECRKMIGGVDKDGD 159
> pfa:PF13_0211 calcium-dependent protein kinase, putative (EC:2.7.11.17);
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=568
Score = 61.2 bits (147), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 43/135 (31%), Positives = 61/135 (45%), Gaps = 16/135 (11%)
Query 16 ELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLEAVDFDK 75
+L K F+ D NGDG L E+ + LK D + + E+ +L+ +D D
Sbjct 429 KLKKTFEAFDHNGDGVLTISEIFQC------LKVND------NEFDRELYFLLKQLDTDG 476
Query 76 NGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATIFGVS----ELDS 131
NG I+Y+EF+ +D AF +FD DG G I+ EL I S
Sbjct 477 NGLIDYTEFLAACLDHSIFQQDVICRNAFNVFDLDGDGVITKDELFKILSFSAVQVSFSK 536
Query 132 EAWRRVLAEVDRNND 146
E ++ EVD NND
Sbjct 537 EIIENLIKEVDSNND 551
Score = 29.3 bits (64), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 25/110 (22%), Positives = 50/110 (45%), Gaps = 27/110 (24%)
Query 2 LYMGSKLTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVE---------------LMK 46
++ K+ NE EL + +++D +G+G +D E + ++ +
Sbjct 450 IFQCLKVNDNEFDRELYFLLKQLDTDGNGLIDYTEFLAACLDHSIFQQDVICRNAFNVFD 509
Query 47 LKGEDV-------SVLDKSAIETE-----VEQVLEAVDFDKNGFIEYSEF 84
L G+ V +L SA++ +E +++ VD + +GFI+Y EF
Sbjct 510 LDGDGVITKDELFKILSFSAVQVSFSKEIIENLIKEVDSNNDGFIDYDEF 559
> cel:C24H10.5 uvt-2; Unidentified Vitellogenin-linked Transcript
family member (uvt-2)
Length=156
Score = 61.2 bits (147), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 47/135 (34%), Positives = 71/135 (52%), Gaps = 18/135 (13%)
Query 15 DELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLEAVDFD 74
D+L IF++ D NGDG + ++EL +M+ G+ S E E++ + +A D D
Sbjct 21 DDLKGIFREFDLNGDGYIQREELR----AVMQKMGQ-------SPTEDELDAMFQAADKD 69
Query 75 KNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATIF---GVSELDS 131
+G I++ EF+ +A LS L+ FE D DG G I+ SEL T F G S L
Sbjct 70 CDGNIDFQEFLVIAKANPLSLS---LKAVFEELDVDGDGYITRSELRTAFQRMGHS-LSD 125
Query 132 EAWRRVLAEVDRNND 146
+ + + VD+NND
Sbjct 126 QDIKAIYRHVDQNND 140
Score = 42.4 bits (98), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 30/115 (26%), Positives = 50/115 (43%), Gaps = 14/115 (12%)
Query 7 KLTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQ 66
K+ + DEL+ +FQ DK+ DG +D QE + + + + ++
Sbjct 49 KMGQSPTEDELDAMFQAADKDCDGNIDFQEFL--------------VIAKANPLSLSLKA 94
Query 67 VLEAVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELA 121
V E +D D +G+I SE T LS Q ++ + D + GKI+ E
Sbjct 95 VFEELDVDGDGYITRSELRTAFQRMGHSLSDQDIKAIYRHVDQNNDGKINFQEFC 149
> tgo:TGME49_042400 calcium-dependent protein kinase, putative
(EC:2.7.11.17); K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=604
Score = 60.8 bits (146), Expect = 1e-09, Method: Composition-based stats.
Identities = 34/111 (30%), Positives = 59/111 (53%), Gaps = 17/111 (15%)
Query 17 LNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIE---TEVEQVLEAVDF 73
L IF +D++GDG L E+ G L KS ++ ++++ ++ VD
Sbjct 435 LKNIFLALDEDGDGTLTINEIRVG--------------LSKSGLKEMPSDLDALMNEVDS 480
Query 74 DKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATIF 124
D +G I+Y+EF+ ++D+R + AF +FD D +G+IS+ ELA +
Sbjct 481 DGSGVIDYTEFIAASLDKRQYIQEDVCWAAFRVFDLDNNGRISADELAQLL 531
Score = 30.4 bits (67), Expect = 2.3, Method: Composition-based stats.
Identities = 30/143 (20%), Positives = 57/143 (39%), Gaps = 18/143 (12%)
Query 6 SKLTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVE 65
SK E +L+ + ++D +G G +D E + ++ + EDV
Sbjct 460 SKSGLKEMPSDLDALMNEVDSDGSGVIDYTEFIAASLDKRQYIQEDV-----------CW 508
Query 66 QVLEAVDFDKNGFIEYSEF--VTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATI 123
D D NG I E + V++D + + ++ + GK+++ E
Sbjct 509 AAFRVFDLDNNGRISADELAQLLVSVDVQNMFPQRE-----GLGQPGAEGKLTAEEKKGA 563
Query 124 FGVSELDSEAWRRVLAEVDRNND 146
+L+ + ++ EVDRN D
Sbjct 564 REQYKLNVLEMKGLIKEVDRNGD 586
Score = 29.6 bits (65), Expect = 3.5, Method: Composition-based stats.
Identities = 10/24 (41%), Positives = 18/24 (75%), Gaps = 0/24 (0%)
Query 16 ELNKIFQKMDKNGDGQLDKQELME 39
E+ + +++D+NGDG++D E ME
Sbjct 573 EMKGLIKEVDRNGDGEIDFDEFME 596
> ath:AT3G51850 CPK13; CPK13; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase/ protein tyrosine kinase;
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=528
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 37/116 (31%), Positives = 61/116 (52%), Gaps = 11/116 (9%)
Query 6 SKLTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVE 65
++ + EE +++ +F KMD + DG + +EL G D S E+EV+
Sbjct 349 AEFLSTEEVEDIKVMFNKMDTDNDGIVSIEELKAGL--------RDFST---QLAESEVQ 397
Query 66 QVLEAVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELA 121
++EAVD G ++Y EFV V++ + + + + L +AF FD DG+G I EL
Sbjct 398 MLIEAVDTKGKGTLDYGEFVAVSLHLQKVANDEHLRKAFSYFDKDGNGYILPQELC 453
Score = 31.6 bits (70), Expect = 0.93, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 33/73 (45%), Gaps = 11/73 (15%)
Query 15 DELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLEAVDFD 74
+ L K F DK+G+G + QEL + +K G D V + + VD D
Sbjct 430 EHLRKAFSYFDKDGNGYILPQELCDA----LKEDGGDDCV-------DVANDIFQEVDTD 478
Query 75 KNGFIEYSEFVTV 87
K+G I Y EF +
Sbjct 479 KDGRISYEEFAAM 491
> ath:AT4G35310 CPK5; CPK5 (calmodulin-domain protein kinase 5);
ATP binding / calcium ion binding / calmodulin-dependent
protein kinase/ kinase/ protein kinase/ protein serine/threonine
kinase; K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=556
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 42/137 (30%), Positives = 66/137 (48%), Gaps = 11/137 (8%)
Query 10 TNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLE 69
+ EE L ++FQ MD + G + EL G ++ G + +TE+ +++
Sbjct 396 SEEEIAGLREMFQAMDTDNSGAITFDELKAG----LRKYGSTLK-------DTEIHDLMD 444
Query 70 AVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATIFGVSEL 129
A D D +G I+YSEF+ + L + L AF+ FD DGSG I+ EL +
Sbjct 445 AADVDNSGTIDYSEFIAATIHLNKLEREEHLVAAFQYFDKDGSGFITIDELQQACVEHGM 504
Query 130 DSEAWRRVLAEVDRNND 146
++ EVD+NND
Sbjct 505 ADVFLEDIIKEVDQNND 521
Score = 30.0 bits (66), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 24/89 (26%), Positives = 38/89 (42%), Gaps = 19/89 (21%)
Query 12 EETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLEAV 71
E + L FQ DK+G G + EL + VE + +E +++ V
Sbjct 470 EREEHLVAAFQYFDKDGSGFITIDELQQACVE-------------HGMADVFLEDIIKEV 516
Query 72 DFDKNGFIEYSEFV------TVAMDRRTL 94
D + +G I+Y EFV + RRT+
Sbjct 517 DQNNDGKIDYGEFVEMMQKGNAGVGRRTM 545
> ath:AT4G23650 CDPK6; CDPK6 (CALCIUM-DEPENDENT PROTEIN KINASE
6); ATP binding / calcium ion binding / calmodulin-dependent
protein kinase/ kinase/ protein kinase/ protein serine/threonine
kinase; K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=529
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 42/138 (30%), Positives = 70/138 (50%), Gaps = 12/138 (8%)
Query 10 TNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLE 69
+ EE L ++F+ +D + +G + +EL G +L G +S E E+ Q++E
Sbjct 377 SEEEIIGLKEMFKSLDTDNNGIVTLEELRTGLPKL----GSKIS-------EAEIRQLME 425
Query 70 AVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATIFGVSEL 129
A D D +G I+Y EF++ M + L AF+ FD D SG I+ EL +
Sbjct 426 AADMDGDGSIDYLEFISATMHMNRIEREDHLYTAFQFFDNDNSGYITMEELELAMKKYNM 485
Query 130 -DSEAWRRVLAEVDRNND 146
D ++ + ++AEVD + D
Sbjct 486 GDDKSIKEIIAEVDTDRD 503
> ath:AT4G38230 CPK26; CPK26; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase;
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=340
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 42/137 (30%), Positives = 65/137 (47%), Gaps = 11/137 (8%)
Query 10 TNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLE 69
+ EE L ++F+ MD + G + EL G ++ G + +TE+ ++E
Sbjct 179 SEEEIAGLKEMFKAMDTDNSGAITFDELKAG----LRRYGSTLK-------DTEIRDLME 227
Query 70 AVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATIFGVSEL 129
A D DK+G I+Y EF+ + L + L AF FD DGSG I+ EL +
Sbjct 228 AADIDKSGTIDYGEFIAATIHLNKLEREEHLLSAFRYFDKDGSGYITIDELQHACAEQGM 287
Query 130 DSEAWRRVLAEVDRNND 146
V+ EVD++ND
Sbjct 288 SDVFLEDVIKEVDQDND 304
Score = 30.0 bits (66), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 25/91 (27%), Positives = 40/91 (43%), Gaps = 15/91 (16%)
Query 7 KLTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQ 66
L E + L F+ DK+G G + EL E + + +E
Sbjct 248 HLNKLEREEHLLSAFRYFDKDGSGYITIDELQHACAE-------------QGMSDVFLED 294
Query 67 VLEAVDFDKNGFIEYSEFVTVAMDRRTLLSR 97
V++ VD D +G I+Y EF VAM ++ ++ R
Sbjct 295 VIKEVDQDNDGRIDYGEF--VAMMQKGIVGR 323
> ath:AT1G32250 calmodulin, putative; K13448 calcium-binding protein
CML
Length=166
Score = 59.3 bits (142), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 39/123 (31%), Positives = 61/123 (49%), Gaps = 18/123 (14%)
Query 6 SKLTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVE 65
SK E+ +EL +IF+ D+N DG L + EL G + L + E
Sbjct 6 SKKLDEEQINELREIFRSFDRNKDGSLTQLEL-----------GSLLRALGVKPSPDQFE 54
Query 66 QVLEAVDFDKNGFIEYSEFVTV-------AMDRRTLLSRQRLERAFEMFDTDGSGKISSS 118
+++ D NG +E+ EFV + R T + ++L R F +FDTDG+G I+++
Sbjct 55 TLIDKADTKSNGLVEFPEFVALVSPELLSPAKRTTPYTEEQLLRLFRIFDTDGNGFITAA 114
Query 119 ELA 121
ELA
Sbjct 115 ELA 117
> ath:AT2G17290 CPK6; CPK6 (CALCIUM-DEPENDENT PROTEIN KINASE 6);
ATP binding / calcium ion binding / calmodulin-dependent
protein kinase/ kinase/ protein kinase/ protein serine/threonine
kinase; K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=544
Score = 57.8 bits (138), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 41/137 (29%), Positives = 65/137 (47%), Gaps = 11/137 (8%)
Query 10 TNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLE 69
+ EE L +F+ MD + G + EL G ++ G + +TE+ ++E
Sbjct 384 SEEEIAGLRAMFEAMDTDNSGAITFDELKAG----LRRYGSTLK-------DTEIRDLME 432
Query 70 AVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATIFGVSEL 129
A D D +G I+YSEF+ + L + L AF+ FD DGSG I+ EL +
Sbjct 433 AADVDNSGTIDYSEFIAATIHLNKLEREEHLVSAFQYFDKDGSGYITIDELQQSCIEHGM 492
Query 130 DSEAWRRVLAEVDRNND 146
++ EVD++ND
Sbjct 493 TDVFLEDIIKEVDQDND 509
Score = 32.7 bits (73), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 24/89 (26%), Positives = 40/89 (44%), Gaps = 19/89 (21%)
Query 12 EETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLEAV 71
E + L FQ DK+G G + EL + S ++ + +E +++ V
Sbjct 458 EREEHLVSAFQYFDKDGSGYITIDELQQ-------------SCIEHGMTDVFLEDIIKEV 504
Query 72 DFDKNGFIEYSEFVTV------AMDRRTL 94
D D +G I+Y EFV + + RRT+
Sbjct 505 DQDNDGRIDYEEFVAMMQKGNAGVGRRTM 533
> dre:100150680 calmodulin 2-like; K02183 calmodulin
Length=229
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 37/116 (31%), Positives = 61/116 (52%), Gaps = 12/116 (10%)
Query 10 TNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLE 69
T E+ E + F DK+GDG + +EL G + L ++ E E++ ++
Sbjct 86 TEEQIAEFKEAFSLFDKDGDGTITTKEL-----------GTVMRSLGQNPTEAELQDMIN 134
Query 70 AVDFDKNGFIEYSEFVTV-AMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATIF 124
VD D NG I++ EF+T+ A + S + + AF +FD DG+G IS++EL +
Sbjct 135 EVDADGNGTIDFPEFLTMMARKMKDTDSEEEIREAFRVFDKDGNGYISAAELRHVM 190
Score = 33.1 bits (74), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 23/85 (27%), Positives = 42/85 (49%), Gaps = 11/85 (12%)
Query 1 LLYMGSKLTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAI 60
L M K+ + +E+ + F+ DK+G+G + EL +M GE ++
Sbjct 150 LTMMARKMKDTDSEEEIREAFRVFDKDGNGYISAAELR----HVMTNLGEKLT------- 198
Query 61 ETEVEQVLEAVDFDKNGFIEYSEFV 85
+ EV++++ D D +G + Y EFV
Sbjct 199 DEEVDEMIREADIDGDGQVNYEEFV 223
Score = 33.1 bits (74), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 27/113 (23%), Positives = 49/113 (43%), Gaps = 10/113 (8%)
Query 8 LTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQV 67
L N EL + ++D +G+G +D E ++ +M K +D E E+ +
Sbjct 120 LGQNPTEAELQDMINEVDADGNGTIDFPE----FLTMMARKMKDTDS------EEEIREA 169
Query 68 LEAVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSEL 120
D D NG+I +E V + L+ + ++ D DG G+++ E
Sbjct 170 FRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEF 222
> tgo:TGME49_037890 calcium-dependent protein kinase, putative
(EC:2.7.11.17)
Length=1171
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 35/135 (25%), Positives = 64/135 (47%), Gaps = 13/135 (9%)
Query 12 EETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLEAV 71
++ D+L ++F+K+D + G + ++ V + + + E ++ + +
Sbjct 961 QQLDDLERLFRKIDIDNSGCIKMDRMVAVLVTFLDVPRD------------EALRIFQRI 1008
Query 72 DFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATIFGVSELDS 131
D + I YSEF+ + R L++Q + AFE FD D SG IS L + G S DS
Sbjct 1009 DQTRAEEINYSEFLAATLQTRIALNQQLIREAFERFDVDNSGHISLENLRYVLGDS-YDS 1067
Query 132 EAWRRVLAEVDRNND 146
+ +L + DR +
Sbjct 1068 LSVEEILRQCDRKQN 1082
Score = 29.6 bits (65), Expect = 3.6, Method: Composition-based stats.
Identities = 23/97 (23%), Positives = 39/97 (40%), Gaps = 30/97 (30%)
Query 15 DELNKIFQKMDKNGDGQLDKQELM--------------------------EGYVELMKLK 48
DE +IFQ++D+ +++ E + G++ L L+
Sbjct 999 DEALRIFQRIDQTRAEEINYSEFLAATLQTRIALNQQLIREAFERFDVDNSGHISLENLR 1058
Query 49 GEDVSVLDKSAIETEVEQVLEAVDFDKNGFIEYSEFV 85
VL S VE++L D +NG IE+ EF+
Sbjct 1059 ----YVLGDSYDSLSVEEILRQCDRKQNGVIEFDEFM 1091
> dre:406660 calm1a, CaMbeta, sb:cb617, wu:fb08d09, wu:fb69c08,
wu:fj34a08, zgc:63926; calmodulin 1a; K02183 calmodulin
Length=149
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 37/115 (32%), Positives = 61/115 (53%), Gaps = 12/115 (10%)
Query 10 TNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLE 69
T E+ E + F DK+GDG + +EL G + L ++ E E++ ++
Sbjct 6 TEEQIAEFKEAFSLFDKDGDGTITTKEL-----------GTVMRSLGQNPTEAELQDMIN 54
Query 70 AVDFDKNGFIEYSEFVTV-AMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATI 123
VD D NG I++ EF+T+ A + S + + AF +FD DG+G IS++EL +
Sbjct 55 EVDADGNGTIDFPEFLTMMARKMKDTDSEEEIREAFRVFDKDGNGYISAAELRHV 109
Score = 32.3 bits (72), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 27/113 (23%), Positives = 49/113 (43%), Gaps = 10/113 (8%)
Query 8 LTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQV 67
L N EL + ++D +G+G +D E ++ +M K +D E E+ +
Sbjct 40 LGQNPTEAELQDMINEVDADGNGTIDFPE----FLTMMARKMKDTDS------EEEIREA 89
Query 68 LEAVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSEL 120
D D NG+I +E V + L+ + ++ D DG G+++ E
Sbjct 90 FRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEF 142
Score = 32.0 bits (71), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 23/85 (27%), Positives = 42/85 (49%), Gaps = 11/85 (12%)
Query 1 LLYMGSKLTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAI 60
L M K+ + +E+ + F+ DK+G+G + EL +M GE ++
Sbjct 70 LTMMARKMKDTDSEEEIREAFRVFDKDGNGYISAAELR----HVMTNLGEKLT------- 118
Query 61 ETEVEQVLEAVDFDKNGFIEYSEFV 85
+ EV++++ D D +G + Y EFV
Sbjct 119 DEEVDEMIREADIDGDGQVNYEEFV 143
> dre:192322 calm2b, calm2, cb169, zgc:64036; calmodulin 2b, (phosphorylase
kinase, delta); K02183 calmodulin
Length=149
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 37/115 (32%), Positives = 61/115 (53%), Gaps = 12/115 (10%)
Query 10 TNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLE 69
T E+ E + F DK+GDG + +EL G + L ++ E E++ ++
Sbjct 6 TEEQIAEFKEAFSLFDKDGDGTITTKEL-----------GTVMRSLGQNPTEAELQDMIN 54
Query 70 AVDFDKNGFIEYSEFVTV-AMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATI 123
VD D NG I++ EF+T+ A + S + + AF +FD DG+G IS++EL +
Sbjct 55 EVDADGNGTIDFPEFLTMMARKMKDTDSEEEIREAFRVFDKDGNGYISAAELRHV 109
Score = 32.3 bits (72), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 27/113 (23%), Positives = 49/113 (43%), Gaps = 10/113 (8%)
Query 8 LTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQV 67
L N EL + ++D +G+G +D E ++ +M K +D E E+ +
Sbjct 40 LGQNPTEAELQDMINEVDADGNGTIDFPE----FLTMMARKMKDTDS------EEEIREA 89
Query 68 LEAVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSEL 120
D D NG+I +E V + L+ + ++ D DG G+++ E
Sbjct 90 FRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEF 142
Score = 32.0 bits (71), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 23/85 (27%), Positives = 42/85 (49%), Gaps = 11/85 (12%)
Query 1 LLYMGSKLTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAI 60
L M K+ + +E+ + F+ DK+G+G + EL +M GE ++
Sbjct 70 LTMMARKMKDTDSEEEIREAFRVFDKDGNGYISAAELR----HVMTNLGEKLT------- 118
Query 61 ETEVEQVLEAVDFDKNGFIEYSEFV 85
+ EV++++ D D +G + Y EFV
Sbjct 119 DEEVDEMIREADIDGDGQVNYEEFV 143
> dre:368217 calm1b, CaMbeta-2, zgc:73137; calmodulin 1b; K02183
calmodulin
Length=149
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 37/115 (32%), Positives = 61/115 (53%), Gaps = 12/115 (10%)
Query 10 TNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLE 69
T E+ E + F DK+GDG + +EL G + L ++ E E++ ++
Sbjct 6 TEEQIAEFKEAFSLFDKDGDGTITTKEL-----------GTVMRSLGQNPTEAELQDMIN 54
Query 70 AVDFDKNGFIEYSEFVTV-AMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATI 123
VD D NG I++ EF+T+ A + S + + AF +FD DG+G IS++EL +
Sbjct 55 EVDADGNGTIDFPEFLTMMARKMKDTDSEEEIREAFRVFDKDGNGYISAAELRHV 109
Score = 32.3 bits (72), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 27/113 (23%), Positives = 49/113 (43%), Gaps = 10/113 (8%)
Query 8 LTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQV 67
L N EL + ++D +G+G +D E ++ +M K +D E E+ +
Sbjct 40 LGQNPTEAELQDMINEVDADGNGTIDFPE----FLTMMARKMKDTDS------EEEIREA 89
Query 68 LEAVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSEL 120
D D NG+I +E V + L+ + ++ D DG G+++ E
Sbjct 90 FRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEF 142
Score = 32.0 bits (71), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 23/85 (27%), Positives = 42/85 (49%), Gaps = 11/85 (12%)
Query 1 LLYMGSKLTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAI 60
L M K+ + +E+ + F+ DK+G+G + EL +M GE ++
Sbjct 70 LTMMARKMKDTDSEEEIREAFRVFDKDGNGYISAAELR----HVMTNLGEKLT------- 118
Query 61 ETEVEQVLEAVDFDKNGFIEYSEFV 85
+ EV++++ D D +G + Y EFV
Sbjct 119 DEEVDEMIREADIDGDGQVNYEEFV 143
> dre:327379 calm3a, CaMgamma, MGC192068, calm2g, wu:fi05b08,
zgc:86728; calmodulin 3a (phosphorylase kinase, delta); K02183
calmodulin
Length=149
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 37/115 (32%), Positives = 61/115 (53%), Gaps = 12/115 (10%)
Query 10 TNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLE 69
T E+ E + F DK+GDG + +EL G + L ++ E E++ ++
Sbjct 6 TEEQIAEFKEAFSLFDKDGDGTITTKEL-----------GTVMRSLGQNPTEAELQDMIN 54
Query 70 AVDFDKNGFIEYSEFVTV-AMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATI 123
VD D NG I++ EF+T+ A + S + + AF +FD DG+G IS++EL +
Sbjct 55 EVDADGNGTIDFPEFLTMMARKMKDTDSEEEIREAFRVFDKDGNGYISAAELRHV 109
Score = 32.3 bits (72), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 27/113 (23%), Positives = 49/113 (43%), Gaps = 10/113 (8%)
Query 8 LTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQV 67
L N EL + ++D +G+G +D E ++ +M K +D E E+ +
Sbjct 40 LGQNPTEAELQDMINEVDADGNGTIDFPE----FLTMMARKMKDTDS------EEEIREA 89
Query 68 LEAVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSEL 120
D D NG+I +E V + L+ + ++ D DG G+++ E
Sbjct 90 FRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEF 142
Score = 32.0 bits (71), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 23/85 (27%), Positives = 42/85 (49%), Gaps = 11/85 (12%)
Query 1 LLYMGSKLTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAI 60
L M K+ + +E+ + F+ DK+G+G + EL +M GE ++
Sbjct 70 LTMMARKMKDTDSEEEIREAFRVFDKDGNGYISAAELR----HVMTNLGEKLT------- 118
Query 61 ETEVEQVLEAVDFDKNGFIEYSEFV 85
+ EV++++ D D +G + Y EFV
Sbjct 119 DEEVDEMIREADIDGDGQVNYEEFV 143
> dre:321808 calm3b, wu:fb36a09, zgc:55591, zgc:76987; calmodulin
3b (phosphorylase kinase, delta) (EC:2.7.11.19); K02183
calmodulin
Length=149
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 37/115 (32%), Positives = 61/115 (53%), Gaps = 12/115 (10%)
Query 10 TNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLE 69
T E+ E + F DK+GDG + +EL G + L ++ E E++ ++
Sbjct 6 TEEQIAEFKEAFSLFDKDGDGTITTKEL-----------GTVMRSLGQNPTEAELQDMIN 54
Query 70 AVDFDKNGFIEYSEFVTV-AMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATI 123
VD D NG I++ EF+T+ A + S + + AF +FD DG+G IS++EL +
Sbjct 55 EVDADGNGTIDFPEFLTMMARKMKDTDSEEEIREAFRVFDKDGNGYISAAELRHV 109
Score = 32.3 bits (72), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 27/113 (23%), Positives = 49/113 (43%), Gaps = 10/113 (8%)
Query 8 LTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQV 67
L N EL + ++D +G+G +D E ++ +M K +D E E+ +
Sbjct 40 LGQNPTEAELQDMINEVDADGNGTIDFPE----FLTMMARKMKDTDS------EEEIREA 89
Query 68 LEAVDFDKNGFIEYSEFVTVAMDRRTLLSRQRLERAFEMFDTDGSGKISSSEL 120
D D NG+I +E V + L+ + ++ D DG G+++ E
Sbjct 90 FRVFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIREADIDGDGQVNYEEF 142
Score = 32.0 bits (71), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 23/85 (27%), Positives = 42/85 (49%), Gaps = 11/85 (12%)
Query 1 LLYMGSKLTTNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAI 60
L M K+ + +E+ + F+ DK+G+G + EL +M GE ++
Sbjct 70 LTMMARKMKDTDSEEEIREAFRVFDKDGNGYISAAELR----HVMTNLGEKLT------- 118
Query 61 ETEVEQVLEAVDFDKNGFIEYSEFV 85
+ EV++++ D D +G + Y EFV
Sbjct 119 DEEVDEMIREADIDGDGQVNYEEFV 143
> dre:336121 calm2a, CaMalpha, calm2d, cb815, wu:fj49d04, zgc:73045;
calmodulin 2a (phosphorylase kinase, delta); K02183 calmodulin
Length=149
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 37/115 (32%), Positives = 61/115 (53%), Gaps = 12/115 (10%)
Query 10 TNEETDELNKIFQKMDKNGDGQLDKQELMEGYVELMKLKGEDVSVLDKSAIETEVEQVLE 69
T E+ E + F DK+GDG + +EL G + L ++ E E++ ++
Sbjct 6 TEEQIAEFKEAFSLFDKDGDGTITTKEL-----------GTVMRSLGQNPTEAELQDMIN 54
Query 70 AVDFDKNGFIEYSEFVTV-AMDRRTLLSRQRLERAFEMFDTDGSGKISSSELATI 123
VD D NG I++ EF+T+ A + S + + AF +FD DG+G IS++EL +
Sbjct 55 EVDADGNGTIDFPEFLTMMARKMKDTDSEEEIREAFRVFDKDGNGYISAAELRHV 109
Lambda K H
0.313 0.131 0.348
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2872883024
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40