bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
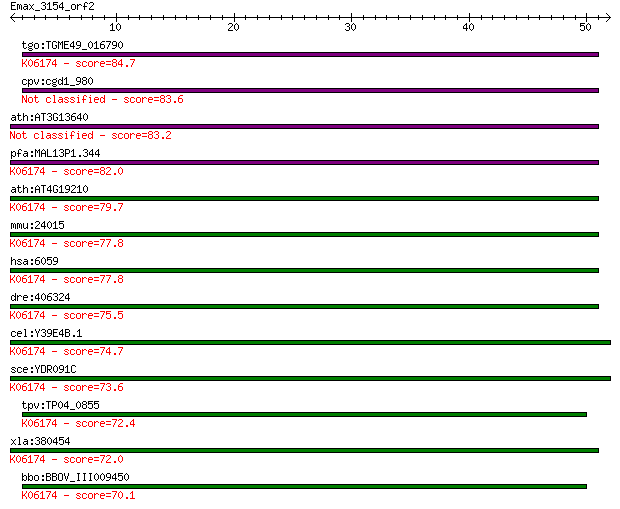
Query= Emax_3154_orf2
Length=51
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_016790 ABC transporter, putative (EC:1.2.7.8 3.6.3.... 84.7 6e-17
cpv:cgd1_980 RNase L inhibitor-like protein 83.6 1e-16
ath:AT3G13640 ATRLI1; ATRLI1; transporter 83.2 2e-16
pfa:MAL13P1.344 RNAse L inhibitor protein, putative; K06174 AT... 82.0 4e-16
ath:AT4G19210 ATRLI2; ATRLI2; transporter; K06174 ATP-binding ... 79.7 2e-15
mmu:24015 Abce1, C79080, Oabp, RLI, RNS41, Rnaseli; ATP-bindin... 77.8 7e-15
hsa:6059 ABCE1, ABC38, OABP, RLI, RNASEL1, RNASELI, RNS4I; ATP... 77.8 7e-15
dre:406324 abce1, MGC56045, wu:fb34c09, wu:fe47b01, wu:fi09g07... 75.5 4e-14
cel:Y39E4B.1 abce-1; ABC transporter, class E family member (a... 74.7 6e-14
sce:YDR091C RLI1; Essential iron-sulfur protein required for r... 73.6 1e-13
tpv:TP04_0855 RNAse L inhibitor; K06174 ATP-binding cassette, ... 72.4 3e-13
xla:380454 abce1, MGC53437; ATP-binding cassette, sub-family E... 72.0 4e-13
bbo:BBOV_III009450 17.m07819; ABC transporter, ATP-binding dom... 70.1 1e-12
> tgo:TGME49_016790 ABC transporter, putative (EC:1.2.7.8 3.6.3.24
3.6.3.25); K06174 ATP-binding cassette, sub-family E, member
1
Length=613
Score = 84.7 bits (208), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 38/49 (77%), Positives = 45/49 (91%), Gaps = 0/49 (0%)
Query 2 LINGMNKFLKVLNITFRRDPTNFRPRINKLDSCKDKEQKLNGNFFVLDD 50
L++GMN+FLK L ITFRRDPTNFRPRINK++S KDKEQKL GN+F+LDD
Sbjct 564 LVSGMNRFLKSLEITFRRDPTNFRPRINKMESVKDKEQKLMGNYFMLDD 612
> cpv:cgd1_980 RNase L inhibitor-like protein
Length=618
Score = 83.6 bits (205), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 36/49 (73%), Positives = 45/49 (91%), Gaps = 0/49 (0%)
Query 2 LINGMNKFLKVLNITFRRDPTNFRPRINKLDSCKDKEQKLNGNFFVLDD 50
L+ GMNKFLK+L I+FRRDP NFRPRINKLDS KDKEQK++GN+F+L++
Sbjct 570 LVTGMNKFLKILEISFRRDPMNFRPRINKLDSVKDKEQKMSGNYFLLEE 618
> ath:AT3G13640 ATRLI1; ATRLI1; transporter
Length=603
Score = 83.2 bits (204), Expect = 2e-16, Method: Composition-based stats.
Identities = 37/50 (74%), Positives = 43/50 (86%), Gaps = 0/50 (0%)
Query 1 SLINGMNKFLKVLNITFRRDPTNFRPRINKLDSCKDKEQKLNGNFFVLDD 50
SL++GMN FL LNITFRRDPTNFRPRINKL+S KDKEQK G+++ LDD
Sbjct 554 SLLSGMNHFLSHLNITFRRDPTNFRPRINKLESIKDKEQKTAGSYYYLDD 603
> pfa:MAL13P1.344 RNAse L inhibitor protein, putative; K06174
ATP-binding cassette, sub-family E, member 1
Length=619
Score = 82.0 bits (201), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 34/50 (68%), Positives = 45/50 (90%), Gaps = 0/50 (0%)
Query 1 SLINGMNKFLKVLNITFRRDPTNFRPRINKLDSCKDKEQKLNGNFFVLDD 50
+L GMNKFLK++++TFRRDP+N+RPRINK DS KDKEQKLNG +F++D+
Sbjct 570 TLAAGMNKFLKIIDVTFRRDPSNYRPRINKYDSVKDKEQKLNGTYFIIDE 619
> ath:AT4G19210 ATRLI2; ATRLI2; transporter; K06174 ATP-binding
cassette, sub-family E, member 1
Length=605
Score = 79.7 bits (195), Expect = 2e-15, Method: Composition-based stats.
Identities = 36/50 (72%), Positives = 43/50 (86%), Gaps = 0/50 (0%)
Query 1 SLINGMNKFLKVLNITFRRDPTNFRPRINKLDSCKDKEQKLNGNFFVLDD 50
SL++GMN FL LNITFRRDPTNFRPRINKL+S KD+EQK G+++ LDD
Sbjct 556 SLLSGMNLFLSHLNITFRRDPTNFRPRINKLESTKDREQKSAGSYYYLDD 605
> mmu:24015 Abce1, C79080, Oabp, RLI, RNS41, Rnaseli; ATP-binding
cassette, sub-family E (OABP), member 1; K06174 ATP-binding
cassette, sub-family E, member 1
Length=599
Score = 77.8 bits (190), Expect = 7e-15, Method: Composition-based stats.
Identities = 35/50 (70%), Positives = 41/50 (82%), Gaps = 0/50 (0%)
Query 1 SLINGMNKFLKVLNITFRRDPTNFRPRINKLDSCKDKEQKLNGNFFVLDD 50
+L+ GMNKFL L ITFRRDP N+RPRINKL+S KD EQK +GN+F LDD
Sbjct 550 TLLAGMNKFLSQLEITFRRDPNNYRPRINKLNSIKDVEQKKSGNYFFLDD 599
> hsa:6059 ABCE1, ABC38, OABP, RLI, RNASEL1, RNASELI, RNS4I; ATP-binding
cassette, sub-family E (OABP), member 1; K06174 ATP-binding
cassette, sub-family E, member 1
Length=599
Score = 77.8 bits (190), Expect = 7e-15, Method: Composition-based stats.
Identities = 35/50 (70%), Positives = 41/50 (82%), Gaps = 0/50 (0%)
Query 1 SLINGMNKFLKVLNITFRRDPTNFRPRINKLDSCKDKEQKLNGNFFVLDD 50
+L+ GMNKFL L ITFRRDP N+RPRINKL+S KD EQK +GN+F LDD
Sbjct 550 TLLAGMNKFLSQLEITFRRDPNNYRPRINKLNSIKDVEQKKSGNYFFLDD 599
> dre:406324 abce1, MGC56045, wu:fb34c09, wu:fe47b01, wu:fi09g07,
zgc:111906, zgc:56045; ATP-binding cassette, sub-family
E (OABP), member 1; K06174 ATP-binding cassette, sub-family
E, member 1
Length=599
Score = 75.5 bits (184), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 36/50 (72%), Positives = 41/50 (82%), Gaps = 0/50 (0%)
Query 1 SLINGMNKFLKVLNITFRRDPTNFRPRINKLDSCKDKEQKLNGNFFVLDD 50
+L+ GMNKFL L ITFRRDP NFRPRINKL+S KD EQK +GN+F LDD
Sbjct 550 TLLAGMNKFLAQLEITFRRDPNNFRPRINKLNSIKDVEQKKSGNYFFLDD 599
> cel:Y39E4B.1 abce-1; ABC transporter, class E family member
(abce-1); K06174 ATP-binding cassette, sub-family E, member
1
Length=610
Score = 74.7 bits (182), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 35/51 (68%), Positives = 42/51 (82%), Gaps = 0/51 (0%)
Query 1 SLINGMNKFLKVLNITFRRDPTNFRPRINKLDSCKDKEQKLNGNFFVLDDN 51
SL+ GMN+FLK+L+ITFRRD +RPRINKLDS KD +QK +G FF LDDN
Sbjct 560 SLLEGMNRFLKMLDITFRRDQETYRPRINKLDSVKDVDQKKSGQFFFLDDN 610
> sce:YDR091C RLI1; Essential iron-sulfur protein required for
ribosome biogenesis and translation initiation; facilitates
binding of a multifactor complex (MFC) of translation initiation
factors to the small ribosomal subunit; predicted ABC
family ATPase; K06174 ATP-binding cassette, sub-family E, member
1
Length=608
Score = 73.6 bits (179), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 35/51 (68%), Positives = 42/51 (82%), Gaps = 0/51 (0%)
Query 1 SLINGMNKFLKVLNITFRRDPTNFRPRINKLDSCKDKEQKLNGNFFVLDDN 51
SL+ G N+FLK LN+TFRRDP +FRPRINKLDS DKEQK +GN+F LD+
Sbjct 556 SLLTGCNRFLKNLNVTFRRDPNSFRPRINKLDSQMDKEQKSSGNYFFLDNT 606
> tpv:TP04_0855 RNAse L inhibitor; K06174 ATP-binding cassette,
sub-family E, member 1
Length=636
Score = 72.4 bits (176), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 32/48 (66%), Positives = 39/48 (81%), Gaps = 0/48 (0%)
Query 2 LINGMNKFLKVLNITFRRDPTNFRPRINKLDSCKDKEQKLNGNFFVLD 49
L G N+FLK L++TFRRDPTN+RPRINK DS KDKEQK +G +F +D
Sbjct 588 LATGFNRFLKSLDVTFRRDPTNYRPRINKYDSVKDKEQKASGLYFTMD 635
> xla:380454 abce1, MGC53437; ATP-binding cassette, sub-family
E (OABP), member 1; K06174 ATP-binding cassette, sub-family
E, member 1
Length=599
Score = 72.0 bits (175), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 33/50 (66%), Positives = 41/50 (82%), Gaps = 0/50 (0%)
Query 1 SLINGMNKFLKVLNITFRRDPTNFRPRINKLDSCKDKEQKLNGNFFVLDD 50
+L+ GMNKFL L ITFRRDP N+RPRINK++S KD +QK +GN+F LDD
Sbjct 550 TLLAGMNKFLSQLEITFRRDPNNYRPRINKMNSIKDVDQKKSGNYFFLDD 599
> bbo:BBOV_III009450 17.m07819; ABC transporter, ATP-binding domain
containing protein; K06174 ATP-binding cassette, sub-family
E, member 1
Length=616
Score = 70.1 bits (170), Expect = 1e-12, Method: Composition-based stats.
Identities = 30/48 (62%), Positives = 39/48 (81%), Gaps = 0/48 (0%)
Query 2 LINGMNKFLKVLNITFRRDPTNFRPRINKLDSCKDKEQKLNGNFFVLD 49
L+ G N+FLK L++TFRRD NFRPRINK DS KDKEQK +G++F ++
Sbjct 568 LVVGFNRFLKSLDVTFRRDQANFRPRINKYDSVKDKEQKASGDYFSVE 615
Lambda K H
0.323 0.142 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2026933688
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40