bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_3147_orf1
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
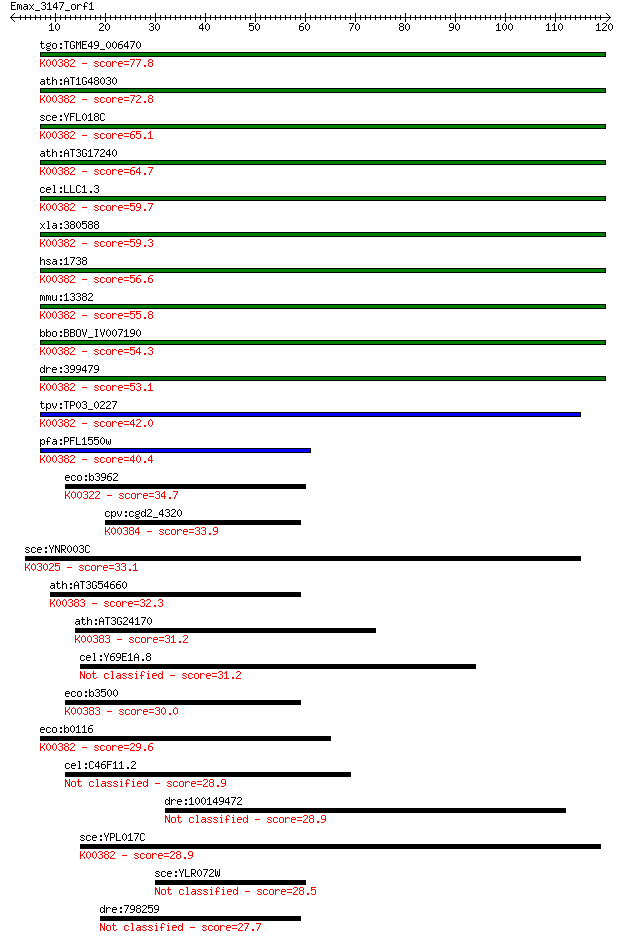
tgo:TGME49_006470 dihydrolipoyl dehydrogenase, putative (EC:1.... 77.8 7e-15
ath:AT1G48030 mtLPD1; mtLPD1 (mitochondrial lipoamide dehydrog... 72.8 3e-13
sce:YFL018C LPD1, HPD1; Dihydrolipoamide dehydrogenase, the li... 65.1 6e-11
ath:AT3G17240 mtLPD2; mtLPD2 (LIPOAMIDE DEHYDROGENASE 2); ATP ... 64.7 6e-11
cel:LLC1.3 hypothetical protein; K00382 dihydrolipoamide dehyd... 59.7 2e-09
xla:380588 dld, MGC68940; dihydrolipoamide dehydrogenase (EC:1... 59.3 3e-09
hsa:1738 DLD, DLDH, E3, GCSL, LAD, PHE3; dihydrolipoamide dehy... 56.6 2e-08
mmu:13382 Dld, AI315664, AI746344; dihydrolipoamide dehydrogen... 55.8 3e-08
bbo:BBOV_IV007190 23.m05858; dihydrolipoamide dehydrogenase (E... 54.3 1e-07
dre:399479 dldh, wu:fb24b05; dihydrolipoamide dehydrogenase (E... 53.1 2e-07
tpv:TP03_0227 dihydrolipoamide dehydrogenase (EC:1.8.1.4); K00... 42.0 5e-04
pfa:PFL1550w lipoamide dehydrogenase (EC:1.8.1.4); K00382 dihy... 40.4 0.001
eco:b3962 sthA, ECK3954, JW5551, sth, udhA; pyridine nucleotid... 34.7 0.072
cpv:cgd2_4320 thioredoxin reductase 1 ; K00384 thioredoxin red... 33.9 0.14
sce:YNR003C RPC34; C34; K03025 DNA-directed RNA polymerase III... 33.1 0.19
ath:AT3G54660 GR; GR (GLUTATHIONE REDUCTASE); ATP binding / gl... 32.3 0.39
ath:AT3G24170 ATGR1; ATGR1 (glutathione-disulfide reductase); ... 31.2 0.83
cel:Y69E1A.8 hypothetical protein 31.2 0.94
eco:b3500 gor, ECK3485, gorA, JW3467; glutathione oxidoreducta... 30.0 1.7
eco:b0116 lpd, dhl, ECK0115, JW0112, lpdA; lipoamide dehydroge... 29.6 2.5
cel:C46F11.2 hypothetical protein 28.9 3.9
dre:100149472 im:7153558 28.9 4.3
sce:YPL017C IRC15; Irc15p; K00382 dihydrolipoamide dehydrogena... 28.9 4.5
sce:YLR072W Protein of unknown function; green fluorescent pro... 28.5 5.1
dre:798259 im:7135991; si:ch1073-179p4.3 27.7 8.7
> tgo:TGME49_006470 dihydrolipoyl dehydrogenase, putative (EC:1.8.1.4);
K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=519
Score = 77.8 bits (190), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 46/113 (40%), Positives = 60/113 (53%), Gaps = 29/113 (25%)
Query 7 LAQGIEGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEAAPLVG 66
L QGIE LF+RN V+Y G G+L D ++VEV P G ++L +IILATGSEA+PL G
Sbjct 141 LTQGIEHLFRRNGVDYYVGEGKLTDSNSVEVTPNGKSEKQRLDAGHIILATGSEASPLPG 200
Query 67 GALEAVTCSRLSTKHLSSRTSRFQRLVFFCLCQVDEQQIVSSTGALALSSVPR 119
+ +DE+ I+SSTGALAL VP+
Sbjct 201 N-----------------------------VVPIDEKVIISSTGALALDKVPK 224
> ath:AT1G48030 mtLPD1; mtLPD1 (mitochondrial lipoamide dehydrogenase
1); ATP binding / dihydrolipoyl dehydrogenase; K00382
dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=507
Score = 72.8 bits (177), Expect = 3e-13, Method: Composition-based stats.
Identities = 46/113 (40%), Positives = 62/113 (54%), Gaps = 32/113 (28%)
Query 7 LAQGIEGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEAAPLVG 66
L +GIEGLFK+NKV Y+KG G+ P+ V V+ I GG+T +K K+II+ATGS+ L G
Sbjct 135 LTRGIEGLFKKNKVTYVKGYGKFISPNEVSVETIDGGNT-IVKGKHIIVATGSDVKSLPG 193
Query 67 GALEAVTCSRLSTKHLSSRTSRFQRLVFFCLCQVDEQQIVSSTGALALSSVPR 119
+DE++IVSSTGAL+LS VP+
Sbjct 194 -------------------------------ITIDEKKIVSSTGALSLSEVPK 215
> sce:YFL018C LPD1, HPD1; Dihydrolipoamide dehydrogenase, the
lipoamide dehydrogenase component (E3) of the pyruvate dehydrogenase
and 2-oxoglutarate dehydrogenase multi-enzyme complexes
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=499
Score = 65.1 bits (157), Expect = 6e-11, Method: Composition-based stats.
Identities = 43/118 (36%), Positives = 57/118 (48%), Gaps = 36/118 (30%)
Query 7 LAQGIEGLFKRNKVEYIKGAGRLADPHTVEVQPIGG--GSTEK---LKTKNIILATGSEA 61
L GIE LFK+NKV Y KG G D + V P+ G G+ ++ L KNII+ATGSE
Sbjct 119 LTGGIELLFKKNKVTYYKGNGSFEDETKIRVTPVDGLEGTVKEDHILDVKNIIVATGSEV 178
Query 62 APLVGGALEAVTCSRLSTKHLSSRTSRFQRLVFFCLCQVDEQQIVSSTGALALSSVPR 119
P G ++DE++IVSSTGAL+L +P+
Sbjct 179 TPFPG-------------------------------IEIDEEKIVSSTGALSLKEIPK 205
> ath:AT3G17240 mtLPD2; mtLPD2 (LIPOAMIDE DEHYDROGENASE 2); ATP
binding / dihydrolipoyl dehydrogenase; K00382 dihydrolipoamide
dehydrogenase [EC:1.8.1.4]
Length=507
Score = 64.7 bits (156), Expect = 6e-11, Method: Composition-based stats.
Identities = 41/113 (36%), Positives = 58/113 (51%), Gaps = 32/113 (28%)
Query 7 LAQGIEGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEAAPLVG 66
L +G+EGLFK+NKV Y+KG G+ P V V I G + +K K+II+ATGS+ L G
Sbjct 135 LTRGVEGLFKKNKVNYVKGYGKFLSPSEVSVDTIDGENV-VVKGKHIIVATGSDVKSLPG 193
Query 67 GALEAVTCSRLSTKHLSSRTSRFQRLVFFCLCQVDEQQIVSSTGALALSSVPR 119
+DE++IVSSTGAL+L+ +P+
Sbjct 194 -------------------------------ITIDEKKIVSSTGALSLTEIPK 215
> cel:LLC1.3 hypothetical protein; K00382 dihydrolipoamide dehydrogenase
[EC:1.8.1.4]
Length=495
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 38/113 (33%), Positives = 55/113 (48%), Gaps = 32/113 (28%)
Query 7 LAQGIEGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEAAPLVG 66
L GI+ LFK NKV +++G + P+TV+ + GS E + +NI++A+GSE P G
Sbjct 120 LTGGIKQLFKANKVGHVEGFATIVGPNTVQAKK-NDGSVETINARNILIASGSEVTPFPG 178
Query 67 GALEAVTCSRLSTKHLSSRTSRFQRLVFFCLCQVDEQQIVSSTGALALSSVPR 119
+DE+QIVSSTGAL+L VP+
Sbjct 179 -------------------------------ITIDEKQIVSSTGALSLGQVPK 200
> xla:380588 dld, MGC68940; dihydrolipoamide dehydrogenase (EC:1.8.1.4);
K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=509
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 40/113 (35%), Positives = 54/113 (47%), Gaps = 32/113 (28%)
Query 7 LAQGIEGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEAAPLVG 66
L GI LFK+NKV +++G G++ + V GST+ + TKNI++ATGSE AP G
Sbjct 134 LTSGIAHLFKQNKVVHVQGFGKITGKNQVTATK-ADGSTQVVNTKNILIATGSEVAPFPG 192
Query 67 GALEAVTCSRLSTKHLSSRTSRFQRLVFFCLCQVDEQQIVSSTGALALSSVPR 119
+DE+ IVSSTGAL+L VP
Sbjct 193 -------------------------------IPIDEETIVSSTGALSLKQVPE 214
> hsa:1738 DLD, DLDH, E3, GCSL, LAD, PHE3; dihydrolipoamide dehydrogenase
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase
[EC:1.8.1.4]
Length=509
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 39/113 (34%), Positives = 51/113 (45%), Gaps = 32/113 (28%)
Query 7 LAQGIEGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEAAPLVG 66
L GI LFK+NKV ++ G G++ + V GG T+ + TKNI++ATGSE P G
Sbjct 134 LTGGIAHLFKQNKVVHVNGYGKITGKNQVTATKADGG-TQVIDTKNILIATGSEVTPFPG 192
Query 67 GALEAVTCSRLSTKHLSSRTSRFQRLVFFCLCQVDEQQIVSSTGALALSSVPR 119
+DE IVSSTGAL+L VP
Sbjct 193 -------------------------------ITIDEDTIVSSTGALSLKKVPE 214
> mmu:13382 Dld, AI315664, AI746344; dihydrolipoamide dehydrogenase
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=509
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 39/113 (34%), Positives = 51/113 (45%), Gaps = 32/113 (28%)
Query 7 LAQGIEGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEAAPLVG 66
L GI LFK+NKV ++ G G++ + V GST+ + TKNI++ATGSE P G
Sbjct 134 LTGGIAHLFKQNKVVHVNGFGKITGKNQVTATK-ADGSTQVIDTKNILVATGSEVTPFPG 192
Query 67 GALEAVTCSRLSTKHLSSRTSRFQRLVFFCLCQVDEQQIVSSTGALALSSVPR 119
+DE IVSSTGAL+L VP
Sbjct 193 -------------------------------ITIDEDTIVSSTGALSLKKVPE 214
> bbo:BBOV_IV007190 23.m05858; dihydrolipoamide dehydrogenase
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=481
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 38/113 (33%), Positives = 54/113 (47%), Gaps = 34/113 (30%)
Query 7 LAQGIEGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEAAPLVG 66
L GI+GLFK+N V+YI G G L + ++++ GG T + KNII+ATGSE G
Sbjct 113 LDAGIKGLFKKNGVDYISGHGTLKSANEIQIE---GGET--VSAKNIIIATGSEVTTFPG 167
Query 67 GALEAVTCSRLSTKHLSSRTSRFQRLVFFCLCQVDEQQIVSSTGALALSSVPR 119
AL ++D ++I+SS AL L VP+
Sbjct 168 DAL-----------------------------KIDGKRIISSDEALVLDEVPK 191
> dre:399479 dldh, wu:fb24b05; dihydrolipoamide dehydrogenase
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=507
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 36/113 (31%), Positives = 51/113 (45%), Gaps = 33/113 (29%)
Query 7 LAQGIEGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEAAPLVG 66
L GI LFK+NKV ++ G G + + V + G + + TKNI++ATGSE P G
Sbjct 133 LTGGIAHLFKQNKVTHVNGFGTITGKNQVTAKTADG--EQVINTKNILIATGSEVTPFPG 190
Query 67 GALEAVTCSRLSTKHLSSRTSRFQRLVFFCLCQVDEQQIVSSTGALALSSVPR 119
++DE +VSSTGAL+L +VP
Sbjct 191 -------------------------------IEIDEDSVVSSTGALSLKNVPE 212
> tpv:TP03_0227 dihydrolipoamide dehydrogenase (EC:1.8.1.4); K00382
dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=499
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 35/108 (32%), Positives = 51/108 (47%), Gaps = 18/108 (16%)
Query 7 LAQGIEGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEAAPLVG 66
L GI GLFK+NK++YI+G + V V GS L K +++ATGSE P
Sbjct 113 LNMGIFGLFKKNKIDYIQGTACFKSQNEVTV-----GSKVLLADK-VVVATGSEVRPFPS 166
Query 67 GALEAVTCSRLSTKHLSSRTSRFQRLVFFCLCQVDEQQIVSSTGALAL 114
+L ++ K+ S T CL +V + +V GA+ L
Sbjct 167 ESL------KVDGKYFLSSTET------LCLDKVPNRLLVIGAGAIGL 202
> pfa:PFL1550w lipoamide dehydrogenase (EC:1.8.1.4); K00382 dihydrolipoamide
dehydrogenase [EC:1.8.1.4]
Length=512
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 21/54 (38%), Positives = 33/54 (61%), Gaps = 2/54 (3%)
Query 7 LAQGIEGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSE 60
L+ GI L+K+N V +I G G L D HTV ++ +K+ + I++ATGS+
Sbjct 116 LSDGINFLYKKNNVNHIIGHGSLVDEHTVLIKT--EKEEKKVTAERIVIATGSK 167
> eco:b3962 sthA, ECK3954, JW5551, sth, udhA; pyridine nucleotide
transhydrogenase, soluble (EC:1.6.1.1); K00322 NAD(P) transhydrogenase
[EC:1.6.1.1]
Length=466
Score = 34.7 bits (78), Expect = 0.072, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 26/48 (54%), Gaps = 1/48 (2%)
Query 12 EGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGS 59
+G ++RN E ++G R D HT+ + GS E L + ++A GS
Sbjct 102 QGFYERNHCEILQGNARFVDEHTLALD-CPDGSVETLTAEKFVIACGS 148
> cpv:cgd2_4320 thioredoxin reductase 1 ; K00384 thioredoxin reductase
(NADPH) [EC:1.8.1.9]
Length=526
Score = 33.9 bits (76), Expect = 0.14, Method: Composition-based stats.
Identities = 16/39 (41%), Positives = 24/39 (61%), Gaps = 2/39 (5%)
Query 20 VEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATG 58
VEYI +L DPH+VE + G + + ++ I+LATG
Sbjct 144 VEYINALAKLIDPHSVEYE--DNGQKKTITSRYILLATG 180
> sce:YNR003C RPC34; C34; K03025 DNA-directed RNA polymerase III
subunit RPC6
Length=317
Score = 33.1 bits (74), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 32/124 (25%), Positives = 57/124 (45%), Gaps = 14/124 (11%)
Query 4 SCVLAQGIEGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIIL-------- 55
S ++++GI LF + +++ G G L D ++ VQ + + KL +N L
Sbjct 20 SQMMSKGIGALFTQQELQKQMGIGSLTDLMSI-VQELLDKNLIKLVKQNDELKFQGVLES 78
Query 56 -----ATGSEAAPLVGGALEAVTCSRLSTKHLSSRTSRFQRLVFFCLCQVDEQQIVSSTG 110
AT S LV +EA + +K + +RT+ Q +V CL ++ Q+ V S
Sbjct 79 EAQKKATMSAEEALVYSYIEASGREGIWSKTIKARTNLHQHVVLKCLKSLESQRYVKSVK 138
Query 111 ALAL 114
++
Sbjct 139 SVKF 142
> ath:AT3G54660 GR; GR (GLUTATHIONE REDUCTASE); ATP binding /
glutathione-disulfide reductase (EC:1.8.1.7); K00383 glutathione
reductase (NADPH) [EC:1.8.1.7]
Length=565
Score = 32.3 bits (72), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 31/54 (57%), Gaps = 10/54 (18%)
Query 9 QGIEGLFK----RNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATG 58
Q + G++K + V+ I+G G++ DPHTV+V + T+NI++A G
Sbjct 186 QRLTGIYKNILSKANVKLIEGRGKVIDPHTVDVD------GKIYTTRNILIAVG 233
> ath:AT3G24170 ATGR1; ATGR1 (glutathione-disulfide reductase);
FAD binding / NADP or NADPH binding / glutathione-disulfide
reductase/ oxidoreductase (EC:1.8.1.7); K00383 glutathione
reductase (NADPH) [EC:1.8.1.7]
Length=499
Score = 31.2 bits (69), Expect = 0.83, Method: Composition-based stats.
Identities = 22/61 (36%), Positives = 32/61 (52%), Gaps = 2/61 (3%)
Query 14 LFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEA-APLVGGALEAV 72
L V+ +G GR+ P+ VEV+ I G K+I++ATGS A P + G A+
Sbjct 133 LLANAAVKLYEGEGRVVGPNEVEVRQIDGTKIS-YTAKHILIATGSRAQKPNIPGHELAI 191
Query 73 T 73
T
Sbjct 192 T 192
> cel:Y69E1A.8 hypothetical protein
Length=364
Score = 31.2 bits (69), Expect = 0.94, Method: Composition-based stats.
Identities = 22/81 (27%), Positives = 39/81 (48%), Gaps = 2/81 (2%)
Query 15 FKRNKVEYIKGAGRLADPHTVEVQPIGGGST--EKLKTKNIILATGSEAAPLVGGALEAV 72
+ +K Y +G+G P +V I G ++ E+ TKN +T +P++ +
Sbjct 191 YSDDKFLYSEGSGLTKSPIDYDVYQIMGSASDIEESPTKNSFHSTLENLSPILKHDMVVA 250
Query 73 TCSRLSTKHLSSRTSRFQRLV 93
+ S +ST+ SS + F LV
Sbjct 251 SSSEISTQSQSSLKAVFHDLV 271
> eco:b3500 gor, ECK3485, gorA, JW3467; glutathione oxidoreductase
(EC:1.8.1.7); K00383 glutathione reductase (NADPH) [EC:1.8.1.7]
Length=450
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 25/47 (53%), Gaps = 6/47 (12%)
Query 12 EGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATG 58
E + +N V+ IKG R D T+EV + E + +I++ATG
Sbjct 100 ENVLGKNNVDVIKGFARFVDAKTLEV------NGETITADHILIATG 140
> eco:b0116 lpd, dhl, ECK0115, JW0112, lpdA; lipoamide dehydrogenase,
E3 component is part of three enzyme complexes (EC:1.8.1.4);
K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=474
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 18/58 (31%), Positives = 29/58 (50%), Gaps = 2/58 (3%)
Query 7 LAQGIEGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEAAPL 64
L G+ G+ K KV+ + G G+ +T+EV+ G + + N I+A GS L
Sbjct 97 LTGGLAGMAKGRKVKVVNGLGKFTGANTLEVEGENGKTV--INFDNAIIAAGSRPIQL 152
> cel:C46F11.2 hypothetical protein
Length=473
Score = 28.9 bits (63), Expect = 3.9, Method: Composition-based stats.
Identities = 20/58 (34%), Positives = 29/58 (50%), Gaps = 7/58 (12%)
Query 12 EGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEAA-PLVGGA 68
E K + VEYI+G A+ TVEV + K + KN ++A G + P + GA
Sbjct 115 ESGLKGSSVEYIRGRATFAEDGTVEV------NGAKYRGKNTLIAVGGKPTIPNIKGA 166
> dre:100149472 im:7153558
Length=699
Score = 28.9 bits (63), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 20/87 (22%), Positives = 39/87 (44%), Gaps = 18/87 (20%)
Query 32 PHTVEVQP---IGGGSTEKLKTKNIILATGSEAAPLVGGALEAVTCSRLSTKHLSSRTSR 88
P + V P + GG+ +KLK N+I G + +G + A+ + SR
Sbjct 87 PQSTGVAPPVTVKGGAMDKLKVDNLISRNGQDTLRTLGLNIRAI-----------EKNSR 135
Query 89 FQRLVFF----CLCQVDEQQIVSSTGA 111
+R F C+ Q+++ +++ + A
Sbjct 136 SEREKIFEEDECIVQINDAELIDKSFA 162
> sce:YPL017C IRC15; Irc15p; K00382 dihydrolipoamide dehydrogenase
[EC:1.8.1.4]
Length=499
Score = 28.9 bits (63), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 26/104 (25%), Positives = 37/104 (35%), Gaps = 32/104 (30%)
Query 15 FKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEAAPLVGGALEAVTC 74
+N V KG DPH VE+ G ++ K I++ATGS G A
Sbjct 119 LSKNNVTVYKGTAAFKDPHHVEIAQ-RGMKPFIVEAKYIVVATGSAVIQCPGVA------ 171
Query 75 SRLSTKHLSSRTSRFQRLVFFCLCQVDEQQIVSSTGALALSSVP 118
+D +I+SS AL+L +P
Sbjct 172 -------------------------IDNDKIISSDKALSLDYIP 190
> sce:YLR072W Protein of unknown function; green fluorescent protein
(GFP)-fusion protein localizes to the cytoplasm in a
punctate pattern; YLR072W is not an esssential gene
Length=693
Score = 28.5 bits (62), Expect = 5.1, Method: Composition-based stats.
Identities = 15/35 (42%), Positives = 18/35 (51%), Gaps = 5/35 (14%)
Query 30 ADPHTVEVQPI-----GGGSTEKLKTKNIILATGS 59
ADPH +V I GGG T NII+AT +
Sbjct 69 ADPHLSDVNSILDNHRGGGETALTSVNNIIMATST 103
> dre:798259 im:7135991; si:ch1073-179p4.3
Length=371
Score = 27.7 bits (60), Expect = 8.7, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 21/40 (52%), Gaps = 1/40 (2%)
Query 19 KVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATG 58
KV+Y+ G L D HTV G + +NI+LATG
Sbjct 144 KVKYLNMKGTLLDKHTVRAVN-AQGKEMTVTARNILLATG 182
Lambda K H
0.317 0.132 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2018002440
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40