bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_3140_orf1
Length=108
Score E
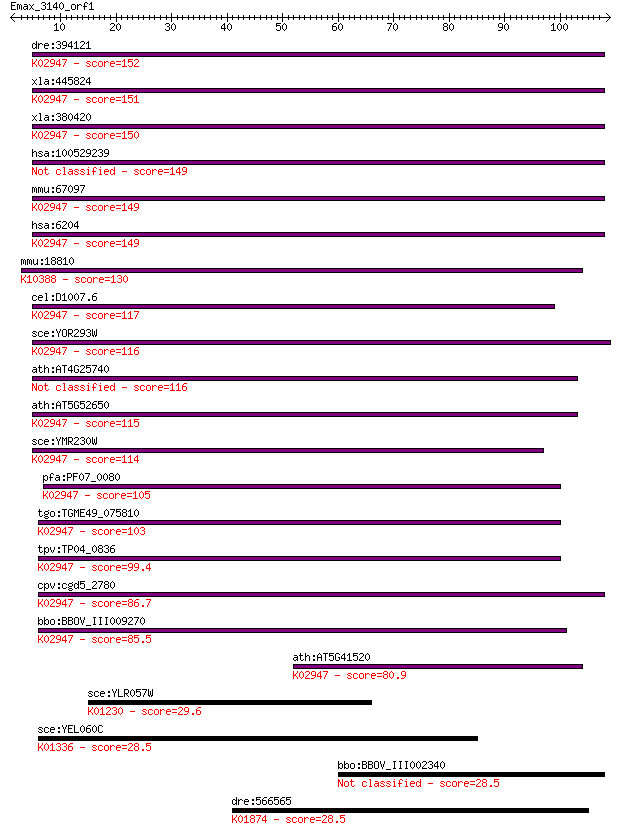
Sequences producing significant alignments: (Bits) Value
dre:394121 rps10, MGC65936, zgc:65936, zgc:85848; ribosomal pr... 152 4e-37
xla:445824 rps10; ribosomal protein S10; K02947 small subunit ... 151 4e-37
xla:380420 rps10, MGC68872; ribosomal protein S10; K02947 smal... 150 9e-37
hsa:100529239 RPS10-NUDT3; RPS10-NUDT3 readthrough 149 2e-36
mmu:67097 Rps10, 2210402A09Rik, MGC107635; ribosomal protein S... 149 2e-36
hsa:6204 RPS10, DBA9, MGC88819; ribosomal protein S10; K02947 ... 149 3e-36
mmu:18810 Plec, AA591047, AU042537, EBS1, PCN, PLTN, Plec1; pl... 130 8e-31
cel:D1007.6 rps-10; Ribosomal Protein, Small subunit family me... 117 7e-27
sce:YOR293W RPS10A; Rps10ap; K02947 small subunit ribosomal pr... 116 2e-26
ath:AT4G25740 40S ribosomal protein S10 (RPS10A) 116 2e-26
ath:AT5G52650 40S ribosomal protein S10 (RPS10C); K02947 small... 115 4e-26
sce:YMR230W RPS10B; Rps10bp; K02947 small subunit ribosomal pr... 114 8e-26
pfa:PF07_0080 40S ribosomal protein S10, putative; K02947 smal... 105 4e-23
tgo:TGME49_075810 ribosomal protein S10, putative ; K02947 sma... 103 2e-22
tpv:TP04_0836 40S ribosomal protein S10; K02947 small subunit ... 99.4 3e-21
cpv:cgd5_2780 40S ribosomal protein S10 ; K02947 small subunit... 86.7 2e-17
bbo:BBOV_III009270 17.m07807; plectin/S10 domain containing pr... 85.5 4e-17
ath:AT5G41520 40S ribosomal protein S10 (RPS10B); K02947 small... 80.9 1e-15
sce:YLR057W Putative protein of unknown function; YLR050C is n... 29.6 2.3
sce:YEL060C PRB1, CVT1; Prb1p (EC:3.4.21.48); K01336 cerevisin... 28.5 4.9
bbo:BBOV_III002340 17.m07228; SmORF 28.5 5.1
dre:566565 Methionyl-tRNA synthetase, mitochondrial-like; K018... 28.5 5.1
> dre:394121 rps10, MGC65936, zgc:65936, zgc:85848; ribosomal
protein S10; K02947 small subunit ribosomal protein S10e
Length=166
Score = 152 bits (383), Expect = 4e-37, Method: Compositional matrix adjust.
Identities = 72/105 (68%), Positives = 90/105 (85%), Gaps = 3/105 (2%)
Query 5 MLMPNKDRLAIFEYLFKEGVMVAEKDFHPKRVHPAL--QKVPNLYVIKALQSLKSKGYVK 62
MLMP K+R+AI+E LFKEGVMVA+KD H + HP L + VPNL+V+KA+QSLKS GYVK
Sbjct 1 MLMPKKNRIAIYELLFKEGVMVAKKDVHLAK-HPELADKNVPNLHVMKAMQSLKSCGYVK 59
Query 63 EQFSWRHYYWYLTNEGIEYLREFLHLPSEIVPSTLKRPIKPESTQ 107
EQF+WRH+YWYLTNEGI+YLR+FLHLP EIVP+TL+R +PE+ +
Sbjct 60 EQFAWRHFYWYLTNEGIQYLRDFLHLPPEIVPATLRRQTRPETAR 104
> xla:445824 rps10; ribosomal protein S10; K02947 small subunit
ribosomal protein S10e
Length=165
Score = 151 bits (382), Expect = 4e-37, Method: Compositional matrix adjust.
Identities = 73/104 (70%), Positives = 90/104 (86%), Gaps = 2/104 (1%)
Query 5 MLMPNKDRLAIFEYLFKEGVMVAEKDFH-PKRVHPALQKVPNLYVIKALQSLKSKGYVKE 63
MLMP KDR+AI+E LFKEGVMVA+KD H PK A + VPNL+V+KA+QSLKS+GYVKE
Sbjct 1 MLMPKKDRIAIYELLFKEGVMVAKKDVHMPKHPELADKNVPNLHVMKAMQSLKSRGYVKE 60
Query 64 QFSWRHYYWYLTNEGIEYLREFLHLPSEIVPSTLKRPIKPESTQ 107
QF+WRH+YWYLTNEGI+YLR+FLHLP EIVP+TL+R +PE+ +
Sbjct 61 QFAWRHFYWYLTNEGIQYLRDFLHLPPEIVPATLRR-SRPETGR 103
> xla:380420 rps10, MGC68872; ribosomal protein S10; K02947 small
subunit ribosomal protein S10e
Length=165
Score = 150 bits (379), Expect = 9e-37, Method: Compositional matrix adjust.
Identities = 75/106 (70%), Positives = 92/106 (86%), Gaps = 6/106 (5%)
Query 5 MLMPNKDRLAIFEYLFKEGVMVAEKDFH-PKRVHPAL--QKVPNLYVIKALQSLKSKGYV 61
MLMP KDR+AI+E LFKEGVMVA+KD H PK HP L + VPNL+V+KA+QSLKS+GYV
Sbjct 1 MLMPKKDRIAIYELLFKEGVMVAKKDVHMPK--HPELVDKNVPNLHVMKAMQSLKSRGYV 58
Query 62 KEQFSWRHYYWYLTNEGIEYLREFLHLPSEIVPSTLKRPIKPESTQ 107
KEQF+WRH+YWYLTNEGI+YLR+FLHLP EIVP+TL+R +PE+ +
Sbjct 59 KEQFAWRHFYWYLTNEGIQYLRDFLHLPPEIVPATLRR-SRPETGR 103
> hsa:100529239 RPS10-NUDT3; RPS10-NUDT3 readthrough
Length=291
Score = 149 bits (376), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 71/104 (68%), Positives = 90/104 (86%), Gaps = 2/104 (1%)
Query 5 MLMPNKDRLAIFEYLFKEGVMVAEKDFH-PKRVHPALQKVPNLYVIKALQSLKSKGYVKE 63
MLMP K+R+AI+E LFKEGVMVA+KD H PK A + VPNL+V+KA+QSLKS+GYVKE
Sbjct 1 MLMPKKNRIAIYELLFKEGVMVAKKDVHMPKHPELADKNVPNLHVMKAMQSLKSRGYVKE 60
Query 64 QFSWRHYYWYLTNEGIEYLREFLHLPSEIVPSTLKRPIKPESTQ 107
QF+WRH+YWYLTNEGI+YLR++LHLP EIVP+TL+R +PE+ +
Sbjct 61 QFAWRHFYWYLTNEGIQYLRDYLHLPPEIVPATLRRS-RPETGR 103
> mmu:67097 Rps10, 2210402A09Rik, MGC107635; ribosomal protein
S10; K02947 small subunit ribosomal protein S10e
Length=165
Score = 149 bits (375), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 71/104 (68%), Positives = 90/104 (86%), Gaps = 2/104 (1%)
Query 5 MLMPNKDRLAIFEYLFKEGVMVAEKDFH-PKRVHPALQKVPNLYVIKALQSLKSKGYVKE 63
MLMP K+R+AI+E LFKEGVMVA+KD H PK A + VPNL+V+KA+QSLKS+GYVKE
Sbjct 1 MLMPKKNRIAIYELLFKEGVMVAKKDVHMPKHPELADKNVPNLHVMKAMQSLKSRGYVKE 60
Query 64 QFSWRHYYWYLTNEGIEYLREFLHLPSEIVPSTLKRPIKPESTQ 107
QF+WRH+YWYLTNEGI+YLR++LHLP EIVP+TL+R +PE+ +
Sbjct 61 QFAWRHFYWYLTNEGIQYLRDYLHLPPEIVPATLRR-SRPETGR 103
> hsa:6204 RPS10, DBA9, MGC88819; ribosomal protein S10; K02947
small subunit ribosomal protein S10e
Length=165
Score = 149 bits (375), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 71/104 (68%), Positives = 90/104 (86%), Gaps = 2/104 (1%)
Query 5 MLMPNKDRLAIFEYLFKEGVMVAEKDFH-PKRVHPALQKVPNLYVIKALQSLKSKGYVKE 63
MLMP K+R+AI+E LFKEGVMVA+KD H PK A + VPNL+V+KA+QSLKS+GYVKE
Sbjct 1 MLMPKKNRIAIYELLFKEGVMVAKKDVHMPKHPELADKNVPNLHVMKAMQSLKSRGYVKE 60
Query 64 QFSWRHYYWYLTNEGIEYLREFLHLPSEIVPSTLKRPIKPESTQ 107
QF+WRH+YWYLTNEGI+YLR++LHLP EIVP+TL+R +PE+ +
Sbjct 61 QFAWRHFYWYLTNEGIQYLRDYLHLPPEIVPATLRR-SRPETGR 103
> mmu:18810 Plec, AA591047, AU042537, EBS1, PCN, PLTN, Plec1;
plectin; K10388 plectin
Length=4691
Score = 130 bits (328), Expect = 8e-31, Method: Compositional matrix adjust.
Identities = 60/102 (58%), Positives = 82/102 (80%), Gaps = 2/102 (1%)
Query 3 ANMLMPNKDRL-AIFEYLFKEGVMVAEKDFHPKRVHPALQKVPNLYVIKALQSLKSKGYV 61
A MLMP DRL AI+E LF+EGVMVA+KD P+ +HP + V NL V++A+ SLK++G V
Sbjct 3 AGMLMP-LDRLRAIYEVLFREGVMVAKKDRRPRSLHPHVPGVTNLQVMRAMASLKARGLV 61
Query 62 KEQFSWRHYYWYLTNEGIEYLREFLHLPSEIVPSTLKRPIKP 103
+E F+W H+YWYLTNEGI++LR++LHLP EIVP++L+R +P
Sbjct 62 RETFAWCHFYWYLTNEGIDHLRQYLHLPPEIVPASLQRVRRP 103
> cel:D1007.6 rps-10; Ribosomal Protein, Small subunit family
member (rps-10); K02947 small subunit ribosomal protein S10e
Length=149
Score = 117 bits (294), Expect = 7e-27, Method: Compositional matrix adjust.
Identities = 57/94 (60%), Positives = 71/94 (75%), Gaps = 1/94 (1%)
Query 5 MLMPNKDRLAIFEYLFKEGVMVAEKDFHPKRVHPALQKVPNLYVIKALQSLKSKGYVKEQ 64
M +P I+EYLF EGV VA+KDF+ K HP ++ V NL VIK L+SL S+ VKEQ
Sbjct 1 MFIPKSHTKLIYEYLFNEGVTVAKKDFNAK-THPNIEGVSNLEVIKTLKSLASRELVKEQ 59
Query 65 FSWRHYYWYLTNEGIEYLREFLHLPSEIVPSTLK 98
F+WRHYYWYLT+ GI YLRE+L LP+EIVP+T+K
Sbjct 60 FAWRHYYWYLTDAGILYLREYLALPAEIVPATIK 93
> sce:YOR293W RPS10A; Rps10ap; K02947 small subunit ribosomal
protein S10e
Length=105
Score = 116 bits (290), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 62/104 (59%), Positives = 77/104 (74%), Gaps = 4/104 (3%)
Query 5 MLMPNKDRLAIFEYLFKEGVMVAEKDFHPKRVHPALQKVPNLYVIKALQSLKSKGYVKEQ 64
MLMP +DR I +YLF+EGV+VA+KDF+ + H + NLYVIKALQSL SKGYVK Q
Sbjct 1 MLMPKEDRNKIHQYLFQEGVVVAKKDFNQAK-HEEID-TKNLYVIKALQSLTSKGYVKTQ 58
Query 65 FSWRHYYWYLTNEGIEYLREFLHLPSEIVPSTLKRPIKPESTQR 108
FSW++YY+ LT EG+EYLRE+L+LP IVP T + P TQR
Sbjct 59 FSWQYYYYTLTEEGVEYLREYLNLPEHIVPGTYIQERNP--TQR 100
> ath:AT4G25740 40S ribosomal protein S10 (RPS10A)
Length=149
Score = 116 bits (290), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 59/99 (59%), Positives = 76/99 (76%), Gaps = 4/99 (4%)
Query 5 MLMPNKDRLAIFEYLFKEGVMVAEKDFH-PKRVHPALQKVPNLYVIKALQSLKSKGYVKE 63
M++ +R I +YLFKEGV A+KDF+ PK HP L VPNL VIK +QS KSK YV+E
Sbjct 1 MIISENNRREICKYLFKEGVCFAKKDFNLPK--HP-LIDVPNLQVIKLMQSFKSKEYVRE 57
Query 64 QFSWRHYYWYLTNEGIEYLREFLHLPSEIVPSTLKRPIK 102
F+W HYYW+LTNEGIE+LR +L+LPS++VP+TLK+ K
Sbjct 58 TFAWMHYYWFLTNEGIEFLRTYLNLPSDVVPATLKKSAK 96
> ath:AT5G52650 40S ribosomal protein S10 (RPS10C); K02947 small
subunit ribosomal protein S10e
Length=179
Score = 115 bits (287), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 57/98 (58%), Positives = 75/98 (76%), Gaps = 2/98 (2%)
Query 5 MLMPNKDRLAIFEYLFKEGVMVAEKDFHPKRVHPALQKVPNLYVIKALQSLKSKGYVKEQ 64
M++ +R I +YLFKEGV A+KDF+ + HP L VPNL VIK +QS KSK YV+E
Sbjct 1 MIISEANRKEICKYLFKEGVCFAKKDFNLAK-HP-LIDVPNLQVIKLMQSFKSKEYVRET 58
Query 65 FSWRHYYWYLTNEGIEYLREFLHLPSEIVPSTLKRPIK 102
F+W HYYW+LTNEGIE+LR +L+LPS++VP+TLK+ K
Sbjct 59 FAWMHYYWFLTNEGIEFLRTYLNLPSDVVPATLKKSAK 96
> sce:YMR230W RPS10B; Rps10bp; K02947 small subunit ribosomal
protein S10e
Length=105
Score = 114 bits (285), Expect = 8e-26, Method: Compositional matrix adjust.
Identities = 57/92 (61%), Positives = 72/92 (78%), Gaps = 2/92 (2%)
Query 5 MLMPNKDRLAIFEYLFKEGVMVAEKDFHPKRVHPALQKVPNLYVIKALQSLKSKGYVKEQ 64
MLMP ++R I +YLF+EGV+VA+KDF+ + H + NLYVIKALQSL SKGYVK Q
Sbjct 1 MLMPKQERNKIHQYLFQEGVVVAKKDFNQAK-HEEI-DTKNLYVIKALQSLTSKGYVKTQ 58
Query 65 FSWRHYYWYLTNEGIEYLREFLHLPSEIVPST 96
FSW++YY+ LT EG+EYLRE+L+LP IVP T
Sbjct 59 FSWQYYYYTLTEEGVEYLREYLNLPEHIVPGT 90
> pfa:PF07_0080 40S ribosomal protein S10, putative; K02947 small
subunit ribosomal protein S10e
Length=137
Score = 105 bits (261), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 52/93 (55%), Positives = 71/93 (76%), Gaps = 2/93 (2%)
Query 7 MPNKDRLAIFEYLFKEGVMVAEKDFHPKRVHPALQKVPNLYVIKALQSLKSKGYVKEQFS 66
+P +++ I+EYLFKEGV+V EKD R HP L VPNL+++ L+SLKS+ YV+E+++
Sbjct 14 IPKQNKKLIYEYLFKEGVIVVEKDAKIPR-HPHLN-VPNLHIMMTLKSLKSRNYVEEKYN 71
Query 67 WRHYYWYLTNEGIEYLREFLHLPSEIVPSTLKR 99
W+H Y+ L NEGIEYLREFLHLP I P+TL +
Sbjct 72 WKHQYFILNNEGIEYLREFLHLPPSIFPATLSK 104
> tgo:TGME49_075810 ribosomal protein S10, putative ; K02947 small
subunit ribosomal protein S10e
Length=152
Score = 103 bits (256), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 50/94 (53%), Positives = 74/94 (78%), Gaps = 2/94 (2%)
Query 6 LMPNKDRLAIFEYLFKEGVMVAEKDFHPKRVHPALQKVPNLYVIKALQSLKSKGYVKEQF 65
L+P +R AI+EYLFKEGV+V +K+ +R HP + VPNL+V+ L+SL++K +V+E+F
Sbjct 10 LIPKANRKAIYEYLFKEGVIVVQKNGKIER-HPEVP-VPNLHVMMTLRSLQTKKFVEEKF 67
Query 66 SWRHYYWYLTNEGIEYLREFLHLPSEIVPSTLKR 99
+W+H Y++LTNEGIE+LR +LHLP + PSTL +
Sbjct 68 NWQHNYYFLTNEGIEFLRTYLHLPPTVFPSTLTK 101
> tpv:TP04_0836 40S ribosomal protein S10; K02947 small subunit
ribosomal protein S10e
Length=114
Score = 99.4 bits (246), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 48/95 (50%), Positives = 72/95 (75%), Gaps = 4/95 (4%)
Query 6 LMPNKDRLAIFEYLFKEGVMVAEKDFHPKR-VHPALQKVPNLYVIKALQSLKSKGYVKEQ 64
L+P ++R+AI+EYL KEGV+V +K+ PK HP + VPNL+V+ ++SLKSK YV+E
Sbjct 10 LIPKENRVAIYEYLIKEGVLVVQKN--PKIPAHPEIS-VPNLHVLMTMRSLKSKNYVEEN 66
Query 65 FSWRHYYWYLTNEGIEYLREFLHLPSEIVPSTLKR 99
+W+H Y+ LT++GIE+LR +LHLP + P+TL +
Sbjct 67 CNWQHLYFTLTDQGIEFLRSYLHLPPTVFPATLTK 101
> cpv:cgd5_2780 40S ribosomal protein S10 ; K02947 small subunit
ribosomal protein S10e
Length=128
Score = 86.7 bits (213), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 46/107 (42%), Positives = 69/107 (64%), Gaps = 7/107 (6%)
Query 6 LMPNKDRLAIFEYLFKEGVMVAEKDFHPKRVHPALQKVPNLYVIKALQSLKSKGYVKEQF 65
L+ K++ AI+ YLFKEG +V K+ + R H L +PNL V+ ++SL SK V E+F
Sbjct 13 LVSKKNQRAIYSYLFKEGAIVVHKNPNDPR-HSELD-IPNLQVMLTMRSLLSKEVVTEKF 70
Query 66 SWRHYYWYLTNEGIEYLREFLHLPSEIVPS-----TLKRPIKPESTQ 107
+W+H Y+ LT+EG+EYLR +L LP+ + P+ T+ RP + E Q
Sbjct 71 TWQHNYYLLTDEGVEYLRRYLDLPASVFPATHTKKTMDRPGRSEGNQ 117
> bbo:BBOV_III009270 17.m07807; plectin/S10 domain containing
protein; K02947 small subunit ribosomal protein S10e
Length=113
Score = 85.5 bits (210), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 43/96 (44%), Positives = 66/96 (68%), Gaps = 3/96 (3%)
Query 6 LMPNKDRLAIFEYLFKEGVMVAEKDFHPKRVHPALQKVPNLYVIKALQSLKSKGYVKEQF 65
++P ++R I+ YL EGV+V +K +HP + VPNL+V+ ++SLKS+ YV E F
Sbjct 10 IIPRENRDMIYRYLINEGVLVCKKSSKIP-LHPEIN-VPNLHVMMVMKSLKSRNYVVEHF 67
Query 66 SWRHYYWYLTNEGIEYLREFLHLPSEIVPST-LKRP 100
+W+H Y+ LT++G+EYLR L+LP + P+T KRP
Sbjct 68 NWQHLYFVLTDQGVEYLRNCLYLPPTVFPATHSKRP 103
> ath:AT5G41520 40S ribosomal protein S10 (RPS10B); K02947 small
subunit ribosomal protein S10e
Length=134
Score = 80.9 bits (198), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 35/52 (67%), Positives = 45/52 (86%), Gaps = 0/52 (0%)
Query 52 LQSLKSKGYVKEQFSWRHYYWYLTNEGIEYLREFLHLPSEIVPSTLKRPIKP 103
+QS KSK YV+E F+W HYYW+LTNEGI++LR +L+LPSEIVP+TLK+ KP
Sbjct 1 MQSFKSKEYVRETFAWMHYYWFLTNEGIDFLRTYLNLPSEIVPATLKKQQKP 52
> sce:YLR057W Putative protein of unknown function; YLR050C is
not an essential gene (EC:3.2.1.-); K01230 mannosyl-oligosaccharide
alpha-1,2-mannosidase [EC:3.2.1.113]
Length=849
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 14/51 (27%), Positives = 29/51 (56%), Gaps = 2/51 (3%)
Query 15 IFEYLFKEGVMVAEKDFHPKRVHPALQKVPNLYVIKALQSLKSKGYVKEQF 65
+FEY +++ D H ++ PA+Q P++ +IK ++ + K + +EQ
Sbjct 133 LFEYSSPSLTSISQNDVH--KIQPAMQLPPDVDMIKQIKDIFMKSWNQEQL 181
> sce:YEL060C PRB1, CVT1; Prb1p (EC:3.4.21.48); K01336 cerevisin
[EC:3.4.21.48]
Length=635
Score = 28.5 bits (62), Expect = 4.9, Method: Composition-based stats.
Identities = 25/92 (27%), Positives = 40/92 (43%), Gaps = 18/92 (19%)
Query 6 LMPNKDRLAIFEYLFKEGVMVAEKDFHPKRVHPA-LQKVPNLYVIKAL--------QSLK 56
++PN+ + +FK G E DFH + V A LQ V NL A S
Sbjct 178 IIPNR-----YIIVFKRGAPQEEIDFHKENVQQAQLQSVENLSAEDAFFISTKDTSLSTS 232
Query 57 SKGYVKEQFS----WRHYYWYLTNEGIEYLRE 84
G +++ F+ + Y Y T E ++ +R+
Sbjct 233 EAGGIQDSFNIDNLFSGYIGYFTQEIVDLIRQ 264
> bbo:BBOV_III002340 17.m07228; SmORF
Length=256
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 23/51 (45%), Gaps = 3/51 (5%)
Query 60 YVKEQFSWRHYYWYLTNEGIEYLREFLHLPSEI---VPSTLKRPIKPESTQ 107
++++ FSW WYL E LPS++ VP PI PE Q
Sbjct 185 HIRDYFSWLGREWYLLPEPQSRAALRKALPSDVAETVPEDCDEPIDPEVEQ 235
> dre:566565 Methionyl-tRNA synthetase, mitochondrial-like; K01874
methionyl-tRNA synthetase [EC:6.1.1.10]
Length=595
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 32/69 (46%), Gaps = 5/69 (7%)
Query 41 QKVPNLYVIKALQSLKSKGYVKEQFSWRHYYWYLTNEGIE---YLREFLHLPSEI--VPS 95
Q NLYV KAL+++ + + F RH W L E +L LH+ E +
Sbjct 457 QHFENLYVYKALEAIGACVRLTNGFVQRHAPWRLDRNNPEEQQWLDTILHVSLECLRIYG 516
Query 96 TLKRPIKPE 104
L +P+ P+
Sbjct 517 ILLQPVVPD 525
Lambda K H
0.320 0.137 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2003209916
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40