bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_3124_orf1
Length=98
Score E
Sequences producing significant alignments: (Bits) Value
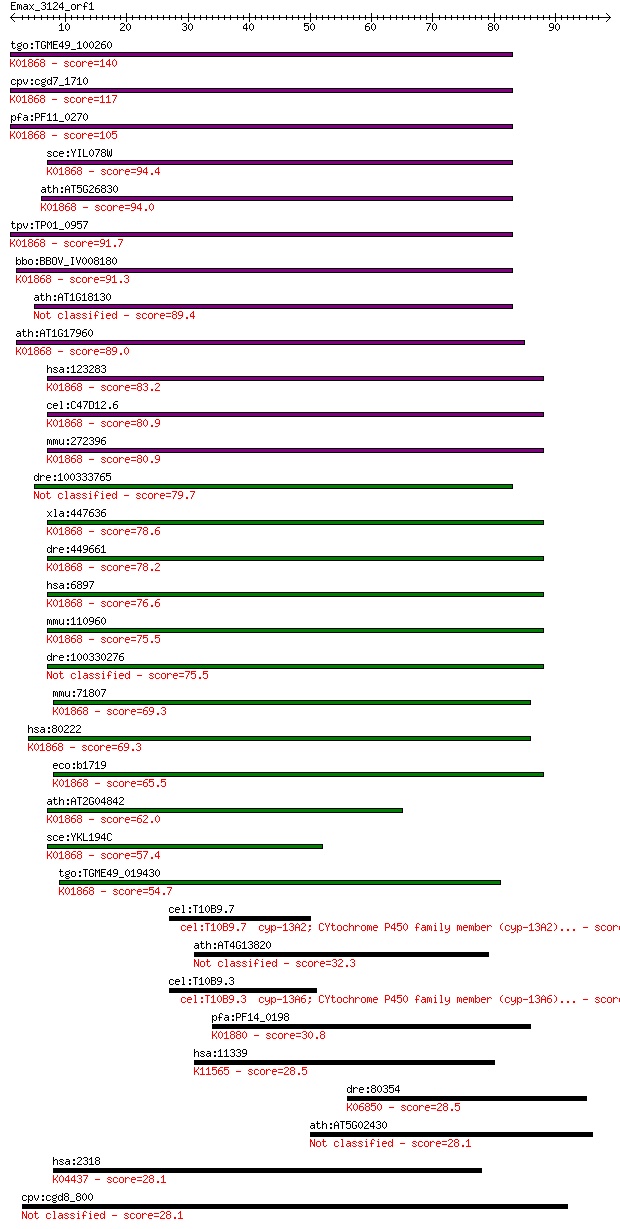
tgo:TGME49_100260 threonyl-tRNA synthetase, putative (EC:6.1.1... 140 1e-33
cpv:cgd7_1710 threonyl-tRNA synthetase (RNA binding domain TGS... 117 8e-27
pfa:PF11_0270 threonine --tRNA ligase, putative; K01868 threon... 105 4e-23
sce:YIL078W THS1; Ths1p (EC:6.1.1.3); K01868 threonyl-tRNA syn... 94.4 8e-20
ath:AT5G26830 threonyl-tRNA synthetase / threonine--tRNA ligas... 94.0 1e-19
tpv:TP01_0957 threonyl-tRNA synthetase; K01868 threonyl-tRNA s... 91.7 5e-19
bbo:BBOV_IV008180 23.m05860; threonyl-tRNA synthetase (EC:6.1.... 91.3 8e-19
ath:AT1G18130 tRNA synthetase-related / tRNA ligase-related (E... 89.4 3e-18
ath:AT1G17960 threonyl-tRNA synthetase, putative / threonine--... 89.0 4e-18
hsa:123283 TARSL2, FLJ25005; threonyl-tRNA synthetase-like 2 (... 83.2 2e-16
cel:C47D12.6 trs-1; Threonyl tRNA Synthetase family member (tr... 80.9 9e-16
mmu:272396 Tarsl2, A530046H20Rik, MGC31414; threonyl-tRNA synt... 80.9 1e-15
dre:100333765 Threonyl-tRNA synthetase, cytoplasmic-like 79.7 2e-15
xla:447636 tars, MGC86352; threonyl-tRNA synthetase (EC:6.1.1.... 78.6 5e-15
dre:449661 tars, ThrRS, wu:fb07c05, wu:fb39c12, zgc:92586; thr... 78.2 6e-15
hsa:6897 TARS, MGC9344, ThrRS; threonyl-tRNA synthetase (EC:6.... 76.6 2e-14
mmu:110960 Tars, D15Wsu59e, ThrRS; threonyl-tRNA synthetase (E... 75.5 3e-14
dre:100330276 threonyl-tRNA synthetase-like 75.5 4e-14
mmu:71807 Tars2, 2610024N01Rik, AI429208, Tarsl1; threonyl-tRN... 69.3 3e-12
hsa:80222 TARS2, FLJ12528, TARSL1; threonyl-tRNA synthetase 2,... 69.3 3e-12
eco:b1719 thrS, ECK1717, JW1709; threonyl-tRNA synthetase (EC:... 65.5 4e-11
ath:AT2G04842 EMB2761 (EMBRYO DEFECTIVE 2761); ATP binding / a... 62.0 4e-10
sce:YKL194C MST1; Mitochondrial threonyl-tRNA synthetase (EC:6... 57.4 1e-08
tgo:TGME49_019430 threonyl-tRNA synthetase, putative (EC:6.1.1... 54.7 6e-08
cel:T10B9.7 cyp-13A2; CYtochrome P450 family member (cyp-13A2)... 33.9 0.15
ath:AT4G13820 disease resistance family protein / LRR family p... 32.3 0.42
cel:T10B9.3 cyp-13A6; CYtochrome P450 family member (cyp-13A6)... 30.8 1.00
pfa:PF14_0198 glycine-tRNA ligase, putative; K01880 glycyl-tRN... 30.8 1.0
hsa:11339 OIP5, 5730547N13Rik, CT86, LINT-25, MIS18B, MIS18bet... 28.5 5.4
dre:80354 slit3, zgc:111911; slit (Drosophila) homolog 3; K068... 28.5 5.7
ath:AT5G02430 WD-40 repeat family protein 28.1 6.4
hsa:2318 FLNC, ABP-280, ABP280A, ABPA, ABPL, FLJ10186, FLN2; f... 28.1 6.8
cpv:cgd8_800 Prp5p C terminal KH. eIF4A-1-family RNA SFII heli... 28.1 7.2
> tgo:TGME49_100260 threonyl-tRNA synthetase, putative (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=844
Score = 140 bits (353), Expect = 1e-33, Method: Composition-based stats.
Identities = 62/82 (75%), Positives = 72/82 (87%), Gaps = 0/82 (0%)
Query 1 LKPGMARPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVA 60
L+PGMARPVIIHRAILGSIERMCAV++EHTGGKLPFWLSP+QCIVL IS+K YA +V
Sbjct 693 LRPGMARPVIIHRAILGSIERMCAVVIEHTGGKLPFWLSPKQCIVLPISDKVNDYAASVR 752
Query 61 DVLQREGYDVGVDVSNNTITRK 82
DVL GY+VG+DVSNNT+ +K
Sbjct 753 DVLHSCGYEVGLDVSNNTVNKK 774
> cpv:cgd7_1710 threonyl-tRNA synthetase (RNA binding domain TGS+HxxxH+tRNA
synthetase) ; K01868 threonyl-tRNA synthetase
[EC:6.1.1.3]
Length=763
Score = 117 bits (293), Expect = 8e-27, Method: Composition-based stats.
Identities = 57/82 (69%), Positives = 63/82 (76%), Gaps = 0/82 (0%)
Query 1 LKPGMARPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVA 60
LK G RPVIIHRAILGS+ERM AV+LEHTGGKLPFWLSPRQ IVLSISEK YA++V
Sbjct 618 LKQGYRRPVIIHRAILGSVERMSAVILEHTGGKLPFWLSPRQAIVLSISEKTVEYAKSVE 677
Query 61 DVLQREGYDVGVDVSNNTITRK 82
L R G+DV D S TI +K
Sbjct 678 RELCRRGFDVSGDYSAATINKK 699
> pfa:PF11_0270 threonine --tRNA ligase, putative; K01868 threonyl-tRNA
synthetase [EC:6.1.1.3]
Length=1013
Score = 105 bits (262), Expect = 4e-23, Method: Composition-based stats.
Identities = 48/82 (58%), Positives = 58/82 (70%), Gaps = 0/82 (0%)
Query 1 LKPGMARPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVA 60
LK G RP+IIHRAILGS+ER A+L+EHT GKLPFWLSPRQ IVL I +K YA V
Sbjct 861 LKKGFERPIIIHRAILGSVERFVAILIEHTAGKLPFWLSPRQAIVLPIGDKYNDYANYVY 920
Query 61 DVLQREGYDVGVDVSNNTITRK 82
+ L +DV +D S NT+ +K
Sbjct 921 ETLHNNMFDVDLDTSVNTLNKK 942
> sce:YIL078W THS1; Ths1p (EC:6.1.1.3); K01868 threonyl-tRNA synthetase
[EC:6.1.1.3]
Length=734
Score = 94.4 bits (233), Expect = 8e-20, Method: Composition-based stats.
Identities = 42/76 (55%), Positives = 54/76 (71%), Gaps = 0/76 (0%)
Query 7 RPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVLQRE 66
RPV+IHRAILGS+ERM A+L EH GK PFWLSPRQ +V+ + K GYAE V + L
Sbjct 594 RPVMIHRAILGSVERMTAILTEHFAGKWPFWLSPRQVLVVPVGVKYQGYAEDVRNKLHDA 653
Query 67 GYDVGVDVSNNTITRK 82
G+ VD++ NT+ +K
Sbjct 654 GFYADVDLTGNTLQKK 669
> ath:AT5G26830 threonyl-tRNA synthetase / threonine--tRNA ligase
(THRRS) (EC:6.1.1.3); K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=709
Score = 94.0 bits (232), Expect = 1e-19, Method: Composition-based stats.
Identities = 42/77 (54%), Positives = 56/77 (72%), Gaps = 0/77 (0%)
Query 6 ARPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVLQR 65
+RPV+IHRA+LGS+ERM A+LLEH GK PFW+SPRQ IV ISEK+ YAE V ++
Sbjct 578 SRPVMIHRAVLGSVERMFAILLEHYKGKWPFWISPRQAIVCPISEKSQQYAEKVQKQIKD 637
Query 66 EGYDVGVDVSNNTITRK 82
G+ V D+++ I +K
Sbjct 638 AGFYVDADLTDRKIDKK 654
> tpv:TP01_0957 threonyl-tRNA synthetase; K01868 threonyl-tRNA
synthetase [EC:6.1.1.3]
Length=779
Score = 91.7 bits (226), Expect = 5e-19, Method: Composition-based stats.
Identities = 43/82 (52%), Positives = 55/82 (67%), Gaps = 0/82 (0%)
Query 1 LKPGMARPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVA 60
++ G RPVIIHRAI GSIER A++LE T GKLPFWLSP Q + + I++ YA +
Sbjct 643 VRCGFNRPVIIHRAIFGSIERFVAIVLEQTKGKLPFWLSPNQLLFIPITDSHLDYALELY 702
Query 61 DVLQREGYDVGVDVSNNTITRK 82
+ GY+V VD SNNTI +K
Sbjct 703 EKFNALGYNVDVDRSNNTINKK 724
> bbo:BBOV_IV008180 23.m05860; threonyl-tRNA synthetase (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=736
Score = 91.3 bits (225), Expect = 8e-19, Method: Composition-based stats.
Identities = 43/81 (53%), Positives = 54/81 (66%), Gaps = 0/81 (0%)
Query 2 KPGMARPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVAD 61
K G +RPVI+HRAI GS+ER A+++EH GKLPFWLSP Q VL IS+K YA +
Sbjct 591 KRGYSRPVIVHRAIFGSLERFIAIVIEHMKGKLPFWLSPTQVAVLPISDKHLDYANKIYR 650
Query 62 VLQREGYDVGVDVSNNTITRK 82
L GY+ VD S NT+ +K
Sbjct 651 KLLHLGYNCYVDSSANTMNKK 671
> ath:AT1G18130 tRNA synthetase-related / tRNA ligase-related
(EC:6.1.1.3)
Length=165
Score = 89.4 bits (220), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 39/78 (50%), Positives = 52/78 (66%), Gaps = 0/78 (0%)
Query 5 MARPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVLQ 64
+ R V+IHRA+L S+ER+ A+LLE GK PFWLSPRQ IV S+SE YA+ V D +
Sbjct 37 IERSVLIHRAVLSSVERLFAILLEQYKGKFPFWLSPRQAIVCSVSEDYRSYADKVRDQIH 96
Query 65 REGYDVGVDVSNNTITRK 82
GY V VD ++ ++ K
Sbjct 97 EAGYYVDVDTTDRNVSDK 114
> ath:AT1G17960 threonyl-tRNA synthetase, putative / threonine--tRNA
ligase, putative; K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=458
Score = 89.0 bits (219), Expect = 4e-18, Method: Composition-based stats.
Identities = 43/83 (51%), Positives = 55/83 (66%), Gaps = 0/83 (0%)
Query 2 KPGMARPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVAD 61
K M PV+IH A+LGSIERM A+LLEH G PFWLSPRQ IV S+SE + YA+ V
Sbjct 319 KNEMEAPVMIHSAVLGSIERMFAILLEHYKGIWPFWLSPRQAIVCSLSEDCSSYAKQVQK 378
Query 62 VLQREGYDVGVDVSNNTITRKSA 84
+ GY V +D S+ ++ +K A
Sbjct 379 QINEVGYYVDIDESDRSLRKKVA 401
> hsa:123283 TARSL2, FLJ25005; threonyl-tRNA synthetase-like 2
(EC:6.1.1.3); K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=802
Score = 83.2 bits (204), Expect = 2e-16, Method: Composition-based stats.
Identities = 39/81 (48%), Positives = 52/81 (64%), Gaps = 0/81 (0%)
Query 7 RPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVLQRE 66
RPVIIHRAILGS+ERM A+L E+ GGK PFWLSPRQ +V+ + YA V+ E
Sbjct 664 RPVIIHRAILGSVERMIAILSENYGGKWPFWLSPRQVMVIPVGPTCEKYALQVSSEFFEE 723
Query 67 GYDVGVDVSNNTITRKSARHS 87
G+ VD+ ++ K R++
Sbjct 724 GFMADVDLDHSCTLNKKIRNA 744
> cel:C47D12.6 trs-1; Threonyl tRNA Synthetase family member (trs-1);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=725
Score = 80.9 bits (198), Expect = 9e-16, Method: Composition-based stats.
Identities = 35/81 (43%), Positives = 50/81 (61%), Gaps = 0/81 (0%)
Query 7 RPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVLQRE 66
RPV+IHRA+LGS+ERM A+L E GGK PFWLSPRQC ++++ E YA V +
Sbjct 584 RPVMIHRAVLGSVERMTAILTESYGGKWPFWLSPRQCKIITVHESVRDYANDVKKQIFEA 643
Query 67 GYDVGVDVSNNTITRKSARHS 87
G+++ + + K R +
Sbjct 644 GFEIEYEENCGDTMNKQVRKA 664
> mmu:272396 Tarsl2, A530046H20Rik, MGC31414; threonyl-tRNA synthetase-like
2 (EC:6.1.1.3); K01868 threonyl-tRNA synthetase
[EC:6.1.1.3]
Length=790
Score = 80.9 bits (198), Expect = 1e-15, Method: Composition-based stats.
Identities = 38/81 (46%), Positives = 51/81 (62%), Gaps = 0/81 (0%)
Query 7 RPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVLQRE 66
RPVIIHRAILGS+ERM A+L E+ GGK P WLSPRQ +V+ + YA V+ E
Sbjct 652 RPVIIHRAILGSVERMIAILSENYGGKWPLWLSPRQVMVIPVGPACENYALQVSKECFEE 711
Query 67 GYDVGVDVSNNTITRKSARHS 87
G+ VD+ ++ K R++
Sbjct 712 GFMADVDLDDSCTLNKKIRNA 732
> dre:100333765 Threonyl-tRNA synthetase, cytoplasmic-like
Length=529
Score = 79.7 bits (195), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 36/79 (45%), Positives = 53/79 (67%), Gaps = 1/79 (1%)
Query 5 MARPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVLQ 64
M RP++IHRA+LGS+ERM A+L E+ GGK PFWLSP Q +V+ + + YA+ V L+
Sbjct 391 MKRPIMIHRAVLGSLERMIAILAENFGGKWPFWLSPAQVMVVPVGGSSEAYAQQVVSRLR 450
Query 65 REGYDVGVDVSN-NTITRK 82
G+ +D + NT+ +K
Sbjct 451 EAGFMADIDDDHGNTLNKK 469
> xla:447636 tars, MGC86352; threonyl-tRNA synthetase (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=721
Score = 78.6 bits (192), Expect = 5e-15, Method: Composition-based stats.
Identities = 37/81 (45%), Positives = 49/81 (60%), Gaps = 0/81 (0%)
Query 7 RPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVLQRE 66
RPVIIHRAILGS+ERM A+L E+ GGK P WLSP+Q +V+ + YA+ V + R
Sbjct 583 RPVIIHRAILGSVERMIAILTENFGGKWPLWLSPQQVMVVPVGPTCDEYAQQVREQFHRA 642
Query 67 GYDVGVDVSNNTITRKSARHS 87
G VD+ K R++
Sbjct 643 GLMTDVDLDPGCTLNKKIRNA 663
> dre:449661 tars, ThrRS, wu:fb07c05, wu:fb39c12, zgc:92586; threonyl-tRNA
synthetase (EC:6.1.1.3); K01868 threonyl-tRNA synthetase
[EC:6.1.1.3]
Length=624
Score = 78.2 bits (191), Expect = 6e-15, Method: Composition-based stats.
Identities = 38/81 (46%), Positives = 48/81 (59%), Gaps = 0/81 (0%)
Query 7 RPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVLQRE 66
RPVIIHRAILGS+ERM A+L E+ GGK P WLSPRQ +V+ I YA+ V +
Sbjct 486 RPVIIHRAILGSVERMIAILTENYGGKWPLWLSPRQVMVVPIGPTCDEYAQRVQKEFHKV 545
Query 67 GYDVGVDVSNNTITRKSARHS 87
G VD+ K R++
Sbjct 546 GLMTDVDLDPGCTLNKKIRNA 566
> hsa:6897 TARS, MGC9344, ThrRS; threonyl-tRNA synthetase (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=723
Score = 76.6 bits (187), Expect = 2e-14, Method: Composition-based stats.
Identities = 35/81 (43%), Positives = 48/81 (59%), Gaps = 0/81 (0%)
Query 7 RPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVLQRE 66
RPVI+HRAILGS+ERM A+L E+ GGK PFWLSPRQ +V+ + YA+ V
Sbjct 585 RPVIVHRAILGSVERMIAILTENYGGKWPFWLSPRQVMVVPVGPTCDEYAQKVRQQFHDA 644
Query 67 GYDVGVDVSNNTITRKSARHS 87
+ +D+ K R++
Sbjct 645 KFMADIDLDPGCTLNKKIRNA 665
> mmu:110960 Tars, D15Wsu59e, ThrRS; threonyl-tRNA synthetase
(EC:6.1.1.3); K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=722
Score = 75.5 bits (184), Expect = 3e-14, Method: Composition-based stats.
Identities = 35/81 (43%), Positives = 47/81 (58%), Gaps = 0/81 (0%)
Query 7 RPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVLQRE 66
RPVI+HRAILGS+ERM A+L E+ GGK PFWLSPRQ +V+ + YA+ V
Sbjct 584 RPVIVHRAILGSVERMIAILTENYGGKWPFWLSPRQVMVVPVGPTCDEYAQKVRQQFHDA 643
Query 67 GYDVGVDVSNNTITRKSARHS 87
+ D+ K R++
Sbjct 644 KFMADTDLDPGCTLNKKIRNA 664
> dre:100330276 threonyl-tRNA synthetase-like
Length=380
Score = 75.5 bits (184), Expect = 4e-14, Method: Composition-based stats.
Identities = 34/81 (41%), Positives = 50/81 (61%), Gaps = 0/81 (0%)
Query 7 RPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVLQRE 66
RPVIIHRAILGS+ERM A+L E+ GK P WLSPRQ +++ ++ YA+ V
Sbjct 242 RPVIIHRAILGSVERMIAILTENYAGKWPLWLSPRQVMLVPVNPSLEEYAKQVCKRFVEA 301
Query 67 GYDVGVDVSNNTITRKSARHS 87
G+ D+ ++ + K R++
Sbjct 302 GFMADADLDSSCLLNKKIRNA 322
> mmu:71807 Tars2, 2610024N01Rik, AI429208, Tarsl1; threonyl-tRNA
synthetase 2, mitochondrial (putative) (EC:6.1.1.3); K01868
threonyl-tRNA synthetase [EC:6.1.1.3]
Length=697
Score = 69.3 bits (168), Expect = 3e-12, Method: Composition-based stats.
Identities = 35/79 (44%), Positives = 47/79 (59%), Gaps = 1/79 (1%)
Query 8 PVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVLQREG 67
PV+IHRA+LGS+ER+ VL E GGK P WLSP Q +V+ + + YA V LQ G
Sbjct 560 PVLIHRAVLGSVERLLGVLAESCGGKWPLWLSPLQVVVIPVRTEQEEYARQVQQCLQAAG 619
Query 68 YDVGVDV-SNNTITRKSAR 85
+D S T++R+ R
Sbjct 620 LVSDLDADSGLTLSRRVRR 638
> hsa:80222 TARS2, FLJ12528, TARSL1; threonyl-tRNA synthetase
2, mitochondrial (putative) (EC:6.1.1.3); K01868 threonyl-tRNA
synthetase [EC:6.1.1.3]
Length=718
Score = 69.3 bits (168), Expect = 3e-12, Method: Composition-based stats.
Identities = 34/83 (40%), Positives = 49/83 (59%), Gaps = 1/83 (1%)
Query 4 GMARPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVL 63
+ RPV+IHRA+LGS+ER+ VL E GGK P WLSP Q +V+ + + YA+ L
Sbjct 577 ALERPVLIHRAVLGSVERLLGVLAESCGGKWPLWLSPFQVVVIPVGSEQEEYAKEAQQSL 636
Query 64 QREGYDVGVDV-SNNTITRKSAR 85
+ G +D S T++R+ R
Sbjct 637 RAAGLVSDLDADSGLTLSRRIRR 659
> eco:b1719 thrS, ECK1717, JW1709; threonyl-tRNA synthetase (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=642
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 30/80 (37%), Positives = 44/80 (55%), Gaps = 0/80 (0%)
Query 8 PVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVLQREG 67
PV+IHRAILGS+ER +L E G P WL+P Q ++++I++ + Y + L G
Sbjct 507 PVMIHRAILGSMERFIGILTEEFAGFFPTWLAPVQVVIMNITDSQSEYVNELTQKLSNAG 566
Query 68 YDVGVDVSNNTITRKSARHS 87
V D+ N I K H+
Sbjct 567 IRVKADLRNEKIGFKIREHT 586
> ath:AT2G04842 EMB2761 (EMBRYO DEFECTIVE 2761); ATP binding /
aminoacyl-tRNA ligase/ ligase, forming aminoacyl-tRNA and related
compounds / nucleotide binding / threonine-tRNA ligase
(EC:6.1.1.3); K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=650
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 26/58 (44%), Positives = 38/58 (65%), Gaps = 0/58 (0%)
Query 7 RPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVADVLQ 64
RP++IHRA+LGS+ER VL+EH G P WLSP Q VL +++ + + V+ L+
Sbjct 519 RPIMIHRAVLGSLERFFGVLIEHYAGDFPLWLSPVQVRVLPVTDNQLEFCKEVSKKLR 576
> sce:YKL194C MST1; Mitochondrial threonyl-tRNA synthetase (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=462
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 21/45 (46%), Positives = 33/45 (73%), Gaps = 0/45 (0%)
Query 7 RPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEK 51
RP++IHRA GSIER A+L++ G+ PFWL+P Q +++ ++ K
Sbjct 314 RPIMIHRATFGSIERFMALLIDSNEGRWPFWLNPYQAVIIPVNTK 358
> tgo:TGME49_019430 threonyl-tRNA synthetase, putative (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=1223
Score = 54.7 bits (130), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 32/79 (40%), Positives = 46/79 (58%), Gaps = 7/79 (8%)
Query 9 VIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKA---AGYAETVADVLQR 65
V+IHRA +GS+ER A+LLE G LP WL+P + +V +S ++ YAE VA VL+
Sbjct 1061 VLIHRAAVGSLERFLALLLELRNGDLPVWLAPVKAVVCPVSVQSREIREYAERVATVLRE 1120
Query 66 ----EGYDVGVDVSNNTIT 80
G+ + S +T T
Sbjct 1121 SVAPSGFSAEKEGSEDTAT 1139
> cel:T10B9.7 cyp-13A2; CYtochrome P450 family member (cyp-13A2);
K00517 [EC:1.14.-.-]
Length=515
Score = 33.9 bits (76), Expect = 0.15, Method: Composition-based stats.
Identities = 13/23 (56%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 27 LEHTGGKLPFWLSPRQCIVLSIS 49
LEH G LPF L PRQCI + ++
Sbjct 444 LEHKGAYLPFGLGPRQCIGMRLA 466
> ath:AT4G13820 disease resistance family protein / LRR family
protein
Length=719
Score = 32.3 bits (72), Expect = 0.42, Method: Composition-based stats.
Identities = 19/49 (38%), Positives = 31/49 (63%), Gaps = 1/49 (2%)
Query 31 GGKLPFWL-SPRQCIVLSISEKAAGYAETVADVLQREGYDVGVDVSNNT 78
GG++P WL S + ++IS+ + E ADV+QR G + +D+S+NT
Sbjct 410 GGQVPQWLWSLPELQYVNISQNSFSGFEGPADVIQRCGELLMLDISSNT 458
> cel:T10B9.3 cyp-13A6; CYtochrome P450 family member (cyp-13A6);
K00517 [EC:1.14.-.-]
Length=518
Score = 30.8 bits (68), Expect = 1.00, Method: Composition-based stats.
Identities = 11/24 (45%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 27 LEHTGGKLPFWLSPRQCIVLSISE 50
LEH G +PF PRQCI + +++
Sbjct 447 LEHNGSYIPFGSGPRQCIGMRLAQ 470
> pfa:PF14_0198 glycine-tRNA ligase, putative; K01880 glycyl-tRNA
synthetase [EC:6.1.1.14]
Length=889
Score = 30.8 bits (68), Expect = 1.0, Method: Composition-based stats.
Identities = 18/54 (33%), Positives = 30/54 (55%), Gaps = 2/54 (3%)
Query 34 LPFWLSPRQCIVLSISEKAA--GYAETVADVLQREGYDVGVDVSNNTITRKSAR 85
LP+ L+P +C +LSIS A Y + + +L + +D S+ +I +K AR
Sbjct 765 LPYKLAPIKCSILSISNNKAFYPYIKQIQMLLNQYNISSKIDNSSVSIGKKYAR 818
> hsa:11339 OIP5, 5730547N13Rik, CT86, LINT-25, MIS18B, MIS18beta,
hMIS18beta; Opa interacting protein 5; K11565 opa-interacting
protein 5
Length=229
Score = 28.5 bits (62), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 27/52 (51%), Gaps = 3/52 (5%)
Query 31 GGKLPFWLSPRQCIVLSISEKAAGYAETVA---DVLQREGYDVGVDVSNNTI 79
G +LP WL P +C V ++ A A++V D+ + G V V+NN +
Sbjct 63 GPQLPSWLQPERCAVFQCAQCHAVLADSVHLAWDLSRSLGAVVFSRVTNNVV 114
> dre:80354 slit3, zgc:111911; slit (Drosophila) homolog 3; K06850
slit 3
Length=1515
Score = 28.5 bits (62), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 21/39 (53%), Gaps = 1/39 (2%)
Query 56 AETVADVLQREGYDVGVDVSNNTITRKSARHSCSNGICC 94
E V D L R+ + S + + R RHSCS+G+CC
Sbjct 1441 GEAVRDYLYRQMAH-KICTSTSRVQRLECRHSCSSGLCC 1478
> ath:AT5G02430 WD-40 repeat family protein
Length=905
Score = 28.1 bits (61), Expect = 6.4, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 24/47 (51%), Gaps = 1/47 (2%)
Query 50 EKAAGYAETVADVLQREGYDVGVDVSN-NTITRKSARHSCSNGICCL 95
EK+ GY+ V D+++RE + +D N+ KS R S G L
Sbjct 296 EKSVGYSSVVKDLMRRENANSTMDFRKFNSYVSKSLRVSKKRGAALL 342
> hsa:2318 FLNC, ABP-280, ABP280A, ABPA, ABPL, FLJ10186, FLN2;
filamin C, gamma; K04437 filamin
Length=2692
Score = 28.1 bits (61), Expect = 6.8, Method: Composition-based stats.
Identities = 23/74 (31%), Positives = 32/74 (43%), Gaps = 5/74 (6%)
Query 8 PVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPR----QCIVLSISEKAAGYAETVADVL 63
P IH G + C V + G L L PR Q V+++ KAAG + V
Sbjct 1625 PFRIHALPTGDASK-CLVTVSIGGHGLGACLGPRIQIGQETVITVDAKAAGEGKVTCTVS 1683
Query 64 QREGYDVGVDVSNN 77
+G ++ VDV N
Sbjct 1684 TPDGAELDVDVVEN 1697
> cpv:cgd8_800 Prp5p C terminal KH. eIF4A-1-family RNA SFII helicase
Length=934
Score = 28.1 bits (61), Expect = 7.2, Method: Composition-based stats.
Identities = 22/92 (23%), Positives = 41/92 (44%), Gaps = 3/92 (3%)
Query 3 PGMARPVIIHRAILGSIERMCAVLLEHTGGKLPFWLSPRQCIVLSISEKAAGYAETVA-- 60
P + R I+H+ I+ +I C V+ + G +P S + ISEK Y E
Sbjct 833 PTLIRQKIVHKDIIKTINENCGVICQVKGIYIPPDSSNPSTNSVHISEKKKLYIEINGPN 892
Query 61 -DVLQREGYDVGVDVSNNTITRKSARHSCSNG 91
+Q+ +++ + +S+ +HS + G
Sbjct 893 HPKVQKAIHEINLIISSIKQQSNVIKHSFTTG 924
Lambda K H
0.323 0.137 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2050913756
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40