bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_3021_orf1
Length=63
Score E
Sequences producing significant alignments: (Bits) Value
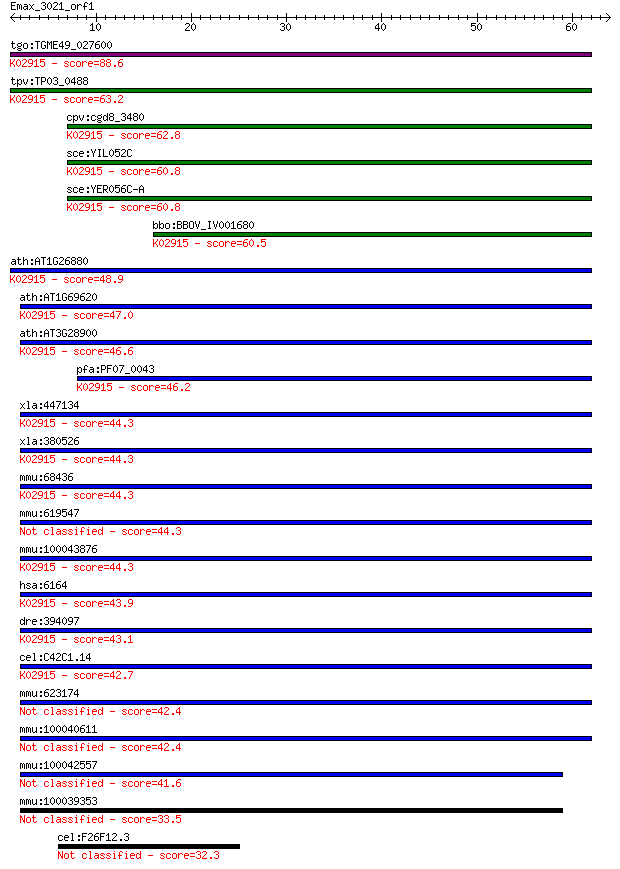
tgo:TGME49_027600 ribosomal protein L34, putative ; K02915 lar... 88.6 4e-18
tpv:TP03_0488 60S ribosomal protein L34a; K02915 large subunit... 63.2 2e-10
cpv:cgd8_3480 60S ribosomal protein L34 ; K02915 large subunit... 62.8 2e-10
sce:YIL052C RPL34B; L34B; K02915 large subunit ribosomal prote... 60.8 1e-09
sce:YER056C-A RPL34A; L34A; K02915 large subunit ribosomal pro... 60.8 1e-09
bbo:BBOV_IV001680 21.m02928; ribosomal protein L34; K02915 lar... 60.5 1e-09
ath:AT1G26880 60S ribosomal protein L34 (RPL34A); K02915 large... 48.9 4e-06
ath:AT1G69620 RPL34; RPL34 (RIBOSOMAL PROTEIN L34); structural... 47.0 1e-05
ath:AT3G28900 60S ribosomal protein L34 (RPL34C); K02915 large... 46.6 2e-05
pfa:PF07_0043 60S ribosomal protein L34-A, putative; K02915 la... 46.2 3e-05
xla:447134 rpl34, MGC85388; ribosomal protein L34; K02915 larg... 44.3 9e-05
xla:380526 rpl34, MGC64481, xl34; ribosomal protein L34; K0291... 44.3 9e-05
mmu:68436 Rpl34, 1100001I22Rik, MGC103270; ribosomal protein L... 44.3 9e-05
mmu:619547 Rpl34-ps1, EG619547, Gm10239, MGC103270, MGC117759;... 44.3 9e-05
mmu:100043876 Gm4705; predicted gene 4705; K02915 large subuni... 44.3 9e-05
hsa:6164 RPL34, MGC111005; ribosomal protein L34; K02915 large... 43.9 1e-04
dre:394097 rpl34, MGC56455, zgc:56455, zgc:77706; ribosomal pr... 43.1 2e-04
cel:C42C1.14 rpl-34; Ribosomal Protein, Large subunit family m... 42.7 3e-04
mmu:623174 Gm6404, EG623174; predicted gene 6404 42.4
mmu:100040611 Gm10154; predicted gene 10154 42.4
mmu:100042557 Gm10086; ribosomal protein L34 pseudogene 41.6 6e-04
mmu:100039353 Gm2178; predicted gene 2178 33.5
cel:F26F12.3 hypothetical protein 32.3 0.37
> tgo:TGME49_027600 ribosomal protein L34, putative ; K02915 large
subunit ribosomal protein L34e
Length=134
Score = 88.6 bits (218), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 44/61 (72%), Positives = 50/61 (81%), Gaps = 0/61 (0%)
Query 1 AKVVYLNVRKQASRQKCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGTRCHSCVRE 60
AKVV+ NV KQ SR KCG C R LPGIPA P + RLLKKRERTV+RAYGG+RCH+CVRE
Sbjct 28 AKVVFHNVGKQPSRPKCGNCHRALPGIPAVAPHRLRLLKKRERTVHRAYGGSRCHACVRE 87
Query 61 K 61
+
Sbjct 88 R 88
> tpv:TP03_0488 60S ribosomal protein L34a; K02915 large subunit
ribosomal protein L34e
Length=138
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 29/61 (47%), Positives = 44/61 (72%), Gaps = 0/61 (0%)
Query 1 AKVVYLNVRKQASRQKCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGTRCHSCVRE 60
A++V N++K R +CG C R L G+ A RP ++ LK+R RTV+R YGG+RCH+CV++
Sbjct 28 ARLVLHNLKKVGKRPRCGDCKRTLAGVSAVRPFLYKNLKRRNRTVSRPYGGSRCHNCVKD 87
Query 61 K 61
+
Sbjct 88 R 88
> cpv:cgd8_3480 60S ribosomal protein L34 ; K02915 large subunit
ribosomal protein L34e
Length=138
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 32/55 (58%), Positives = 40/55 (72%), Gaps = 0/55 (0%)
Query 7 NVRKQASRQKCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGTRCHSCVREK 61
N+ K SR KCG C + L GIPA P + + LKKRERTV RAYGGT+C +CVR++
Sbjct 37 NIGKVYSRPKCGDCKKPLAGIPACAPYEMKHLKKRERTVARAYGGTKCPTCVRQR 91
> sce:YIL052C RPL34B; L34B; K02915 large subunit ribosomal protein
L34e
Length=121
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 28/55 (50%), Positives = 39/55 (70%), Gaps = 0/55 (0%)
Query 7 NVRKQASRQKCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGTRCHSCVREK 61
+V+K A+R KCG CG L GI RP Q+ + K +TV+RAYGG+RC +CV+E+
Sbjct 34 HVKKLATRPKCGDCGSALQGISTLRPRQYATVSKTHKTVSRAYGGSRCANCVKER 88
> sce:YER056C-A RPL34A; L34A; K02915 large subunit ribosomal protein
L34e
Length=121
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 28/55 (50%), Positives = 39/55 (70%), Gaps = 0/55 (0%)
Query 7 NVRKQASRQKCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGTRCHSCVREK 61
+V+K A+R KCG CG L GI RP Q+ + K +TV+RAYGG+RC +CV+E+
Sbjct 34 HVKKLATRPKCGDCGSALQGISTLRPRQYATVSKTHKTVSRAYGGSRCANCVKER 88
> bbo:BBOV_IV001680 21.m02928; ribosomal protein L34; K02915 large
subunit ribosomal protein L34e
Length=157
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 29/46 (63%), Positives = 34/46 (73%), Gaps = 0/46 (0%)
Query 16 KCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGTRCHSCVREK 61
KCG C L GIPA RP +R LKKRER V+RAYGG RCH CV+++
Sbjct 43 KCGDCKCKLAGIPALRPHLYRNLKKRERRVSRAYGGVRCHKCVKDR 88
> ath:AT1G26880 60S ribosomal protein L34 (RPL34A); K02915 large
subunit ribosomal protein L34e
Length=96
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 28/63 (44%), Positives = 38/63 (60%), Gaps = 2/63 (3%)
Query 1 AKVVYLNVRKQASRQKCGGCGRLLPGIPARRPPQFR--LLKKRERTVNRAYGGTRCHSCV 58
K+VY +K+AS KC G+ + GIP RP +++ L + RTVNRAYGG S V
Sbjct 28 GKLVYQTTKKRASGPKCPVTGKRIQGIPHLRPSEYKRSRLSRNRRTVNRAYGGVLSGSAV 87
Query 59 REK 61
RE+
Sbjct 88 RER 90
> ath:AT1G69620 RPL34; RPL34 (RIBOSOMAL PROTEIN L34); structural
constituent of ribosome; K02915 large subunit ribosomal protein
L34e
Length=119
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 27/62 (43%), Positives = 37/62 (59%), Gaps = 2/62 (3%)
Query 2 KVVYLNVRKQASRQKCGGCGRLLPGIPARRPPQFR--LLKKRERTVNRAYGGTRCHSCVR 59
K+ Y +K+AS KC G+ + GIP RP +++ L + RTVNRAYGG S VR
Sbjct 29 KLTYQTTKKRASGPKCPVTGKRIQGIPHLRPTEYKRSRLSRNRRTVNRAYGGVLSGSAVR 88
Query 60 EK 61
E+
Sbjct 89 ER 90
> ath:AT3G28900 60S ribosomal protein L34 (RPL34C); K02915 large
subunit ribosomal protein L34e
Length=120
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 27/62 (43%), Positives = 36/62 (58%), Gaps = 2/62 (3%)
Query 2 KVVYLNVRKQASRQKCGGCGRLLPGIPARRPPQFR--LLKKRERTVNRAYGGTRCHSCVR 59
K+ Y K+AS KC G+ + GIP RP +++ L + ERTVNRAYGG VR
Sbjct 29 KLTYQTTNKRASGPKCPVTGKRIQGIPHLRPAEYKRSRLARNERTVNRAYGGVLSGVAVR 88
Query 60 EK 61
E+
Sbjct 89 ER 90
> pfa:PF07_0043 60S ribosomal protein L34-A, putative; K02915
large subunit ribosomal protein L34e
Length=150
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 22/54 (40%), Positives = 33/54 (61%), Gaps = 0/54 (0%)
Query 8 VRKQASRQKCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGTRCHSCVREK 61
V+K+A + KC C + G+ A RP +++ RTV RAYGG+ C C+RE+
Sbjct 35 VKKKAGKPKCADCKTAIQGVKALRPADNYRARRKNRTVARAYGGSICARCIRER 88
> xla:447134 rpl34, MGC85388; ribosomal protein L34; K02915 large
subunit ribosomal protein L34e
Length=117
Score = 44.3 bits (103), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 26/62 (41%), Positives = 37/62 (59%), Gaps = 2/62 (3%)
Query 2 KVVYLNVRK--QASRQKCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGTRCHSCVR 59
++VYL +K +A + CG C L GI A RP L K ++ V+RAYGG+ C CVR
Sbjct 29 RIVYLYTKKVGKAPKSACGICPGRLRGIRAVRPKVLMRLSKTKKHVSRAYGGSMCAKCVR 88
Query 60 EK 61
++
Sbjct 89 DR 90
> xla:380526 rpl34, MGC64481, xl34; ribosomal protein L34; K02915
large subunit ribosomal protein L34e
Length=117
Score = 44.3 bits (103), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 26/62 (41%), Positives = 37/62 (59%), Gaps = 2/62 (3%)
Query 2 KVVYLNVRK--QASRQKCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGTRCHSCVR 59
++VYL +K +A + CG C L GI A RP L K ++ V+RAYGG+ C CVR
Sbjct 29 RIVYLYTKKVGKAPKSACGICPGRLRGIRAVRPKVLMRLSKTKKHVSRAYGGSMCAKCVR 88
Query 60 EK 61
++
Sbjct 89 DR 90
> mmu:68436 Rpl34, 1100001I22Rik, MGC103270; ribosomal protein
L34; K02915 large subunit ribosomal protein L34e
Length=117
Score = 44.3 bits (103), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 37/62 (59%), Gaps = 2/62 (3%)
Query 2 KVVYLNVRK--QASRQKCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGTRCHSCVR 59
++VYL +K +A + CG C L G+ A RP L K ++ V+RAYGG+ C CVR
Sbjct 29 RIVYLYTKKVGKAPKSACGVCPGRLRGVRAVRPKVLMRLSKTQKHVSRAYGGSMCAKCVR 88
Query 60 EK 61
++
Sbjct 89 DR 90
> mmu:619547 Rpl34-ps1, EG619547, Gm10239, MGC103270, MGC117759;
ribosomal protein L34, pseudogene 1
Length=117
Score = 44.3 bits (103), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 37/62 (59%), Gaps = 2/62 (3%)
Query 2 KVVYLNVRK--QASRQKCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGTRCHSCVR 59
++VYL +K +A + CG C L G+ A RP L K ++ V+RAYGG+ C CVR
Sbjct 29 RIVYLYTKKVGKAPKSACGVCPGRLRGVRAVRPKVLMRLSKTQKHVSRAYGGSMCAKCVR 88
Query 60 EK 61
++
Sbjct 89 DR 90
> mmu:100043876 Gm4705; predicted gene 4705; K02915 large subunit
ribosomal protein L34e
Length=117
Score = 44.3 bits (103), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 37/62 (59%), Gaps = 2/62 (3%)
Query 2 KVVYLNVRK--QASRQKCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGTRCHSCVR 59
++VYL +K +A + CG C L G+ A RP L K ++ V+RAYGG+ C CVR
Sbjct 29 RIVYLYTKKVGKAPKSACGVCPGRLRGVRAVRPKVLMRLSKTQKHVSRAYGGSMCAKCVR 88
Query 60 EK 61
++
Sbjct 89 DR 90
> hsa:6164 RPL34, MGC111005; ribosomal protein L34; K02915 large
subunit ribosomal protein L34e
Length=117
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 37/62 (59%), Gaps = 2/62 (3%)
Query 2 KVVYLNVRK--QASRQKCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGTRCHSCVR 59
++VYL +K +A + CG C L G+ A RP L K ++ V+RAYGG+ C CVR
Sbjct 29 RIVYLYTKKVGKAPKSACGVCPGRLRGVRAVRPKVLMRLSKTKKHVSRAYGGSMCAKCVR 88
Query 60 EK 61
++
Sbjct 89 DR 90
> dre:394097 rpl34, MGC56455, zgc:56455, zgc:77706; ribosomal
protein L34; K02915 large subunit ribosomal protein L34e
Length=117
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 37/62 (59%), Gaps = 2/62 (3%)
Query 2 KVVYLNVRK--QASRQKCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGTRCHSCVR 59
++VYL +K ++ + CG C L GI A RP L K ++ V+RAYGG+ C CVR
Sbjct 29 RIVYLYTKKTGKSPKSACGICPGRLRGIRAVRPQVLMRLSKTKKHVSRAYGGSMCAKCVR 88
Query 60 EK 61
++
Sbjct 89 DR 90
> cel:C42C1.14 rpl-34; Ribosomal Protein, Large subunit family
member (rpl-34); K02915 large subunit ribosomal protein L34e
Length=110
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 25/60 (41%), Positives = 34/60 (56%), Gaps = 0/60 (0%)
Query 2 KVVYLNVRKQASRQKCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGTRCHSCVREK 61
++V ++K+ KC G L GI RP RLLK+ ERTV RAYGG + V+E+
Sbjct 29 RLVVQYIKKRGQIPKCRDTGVKLHGITPARPIALRLLKRNERTVTRAYGGCLSPNAVKER 88
> mmu:623174 Gm6404, EG623174; predicted gene 6404
Length=117
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 36/62 (58%), Gaps = 2/62 (3%)
Query 2 KVVYLNVRK--QASRQKCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGTRCHSCVR 59
++VYL +K +A + CG C L G+ A RP L K ++ V+RAYG + C C+R
Sbjct 29 RIVYLYTKKVGKAPKSACGVCPGRLRGVRAVRPKVLMRLSKTQKQVSRAYGSSMCAKCIR 88
Query 60 EK 61
++
Sbjct 89 DR 90
> mmu:100040611 Gm10154; predicted gene 10154
Length=117
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 24/62 (38%), Positives = 36/62 (58%), Gaps = 2/62 (3%)
Query 2 KVVYLNVRK--QASRQKCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGTRCHSCVR 59
++VYL +K +A + CG C L G+ A RP L K ++ V+RAY G+ C CVR
Sbjct 29 RIVYLYTKKVGKAPKSACGVCPGRLRGVRAVRPKVLMRLSKTQKHVSRAYDGSMCAKCVR 88
Query 60 EK 61
++
Sbjct 89 DR 90
> mmu:100042557 Gm10086; ribosomal protein L34 pseudogene
Length=117
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 24/59 (40%), Positives = 34/59 (57%), Gaps = 2/59 (3%)
Query 2 KVVYLNVRK--QASRQKCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGTRCHSCV 58
++VYL +K +A + CG C L G+ A RP L K ++ V+RAYGG+ C CV
Sbjct 29 RIVYLYTKKVGKAPKSACGVCPGRLRGVRAVRPKVLMRLSKTQKHVSRAYGGSMCAKCV 87
> mmu:100039353 Gm2178; predicted gene 2178
Length=118
Score = 33.5 bits (75), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 22/60 (36%), Positives = 32/60 (53%), Gaps = 3/60 (5%)
Query 2 KVVYLNVRK--QASRQKCGGCGRLLPGIPARRPPQFRL-LKKRERTVNRAYGGTRCHSCV 58
++VYL +K +A + CG C L G+PA R + L K + V+RAY + C CV
Sbjct 29 RIVYLYTKKVGKAPKSACGVCPLRLQGLPAVRARVVLMRLSKTKMQVSRAYSCSMCAKCV 88
> cel:F26F12.3 hypothetical protein
Length=890
Score = 32.3 bits (72), Expect = 0.37, Method: Composition-based stats.
Identities = 12/19 (63%), Positives = 15/19 (78%), Gaps = 0/19 (0%)
Query 6 LNVRKQASRQKCGGCGRLL 24
+ VR QASR +CGGCGR+
Sbjct 708 MEVRDQASRAECGGCGRVF 726
Lambda K H
0.325 0.138 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2056037364
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40