bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_3010_orf1
Length=134
Score E
Sequences producing significant alignments: (Bits) Value
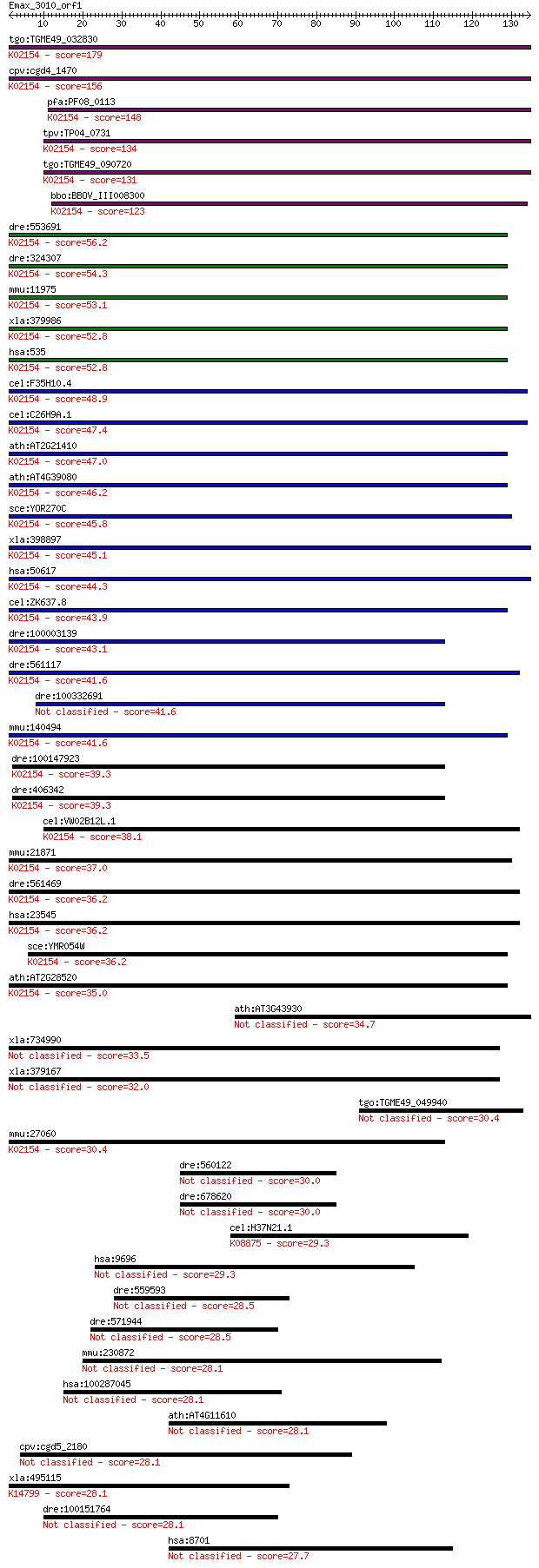
tgo:TGME49_032830 vacuolar proton-translocating ATPase subunit... 179 2e-45
cpv:cgd4_1470 vacuolar proton translocating ATpase with 7 tran... 156 2e-38
pfa:PF08_0113 vacuolar proton translocating ATPase subunit A, ... 148 4e-36
tpv:TP04_0731 vacuolar ATP synthase subunit A (EC:3.6.3.14); K... 134 6e-32
tgo:TGME49_090720 vacuolar proton-translocating ATPase subunit... 131 4e-31
bbo:BBOV_III008300 17.m07725; V-type ATPase 116kDa subunit fam... 123 2e-28
dre:553691 atp6v0a1b, MGC112214, wu:fj37e01, zgc:112214; ATPas... 56.2 3e-08
dre:324307 atp6v0a1a, atp6v0a1, wu:fc25b09, zgc:76965; ATPase,... 54.3 1e-07
mmu:11975 Atp6v0a1, AA959968, ATP6a1, Atp6n1, Atp6n1a, Atpv0a1... 53.1 3e-07
xla:379986 atp6v0a1, MGC52726; ATPase, H+ transporting, lysoso... 52.8 3e-07
hsa:535 ATP6V0A1, ATP6N1, ATP6N1A, DKFZp781J1951, Stv1, VPP1, ... 52.8 3e-07
cel:F35H10.4 vha-5; Vacuolar H ATPase family member (vha-5); K... 48.9 5e-06
cel:C26H9A.1 vha-7; Vacuolar H ATPase family member (vha-7); K... 47.4 1e-05
ath:AT2G21410 VHA-A2; VHA-A2 (VACUOLAR PROTON ATPASE A2); ATPa... 47.0 2e-05
ath:AT4G39080 VHA-A3; VHA-A3 (VACUOLAR PROTON ATPASE A3); ATPa... 46.2 3e-05
sce:YOR270C VPH1; Subunit a of vacuolar-ATPase V0 domain, one ... 45.8 3e-05
xla:398897 atp6v0a4, MGC68661; ATPase, H+ transporting, lysoso... 45.1 6e-05
hsa:50617 ATP6V0A4, A4, ATP6N1B, ATP6N2, MGC130016, MGC130017,... 44.3 9e-05
cel:ZK637.8 unc-32; UNCoordinated family member (unc-32); K021... 43.9 2e-04
dre:100003139 tcirg1, si:dkey-9i23.9; T-cell, immune regulator... 43.1 2e-04
dre:561117 si:ch211-106a19.2 (EC:3.6.3.6); K02154 V-type H+-tr... 41.6 6e-04
dre:100332691 atp6v0a1; ATPase, H+ transporting, lysosomal V0 ... 41.6 6e-04
mmu:140494 Atp6v0a4, Atp6n1b, a4; ATPase, H+ transporting, lys... 41.6 8e-04
dre:100147923 T-cell immune regulator 1-like; K02154 V-type H+... 39.3 0.003
dre:406342 tcirg1, wu:fb44e06, wu:fi46h07; zgc:55891; K02154 V... 39.3 0.004
cel:VW02B12L.1 vha-6; Vacuolar H ATPase family member (vha-6);... 38.1 0.007
mmu:21871 Atp6v0a2, 8430408C20Rik, AI385560, ATP6a2, AW489264,... 37.0 0.019
dre:561469 ATPase, H+ transporting, lysosomal V0 subunit a1-li... 36.2 0.026
hsa:23545 ATP6V0A2, A2, ARCL, ATP6A2, ATP6N1D, J6B7, RTF, STV1... 36.2 0.027
sce:YMR054W STV1; Stv1p (EC:3.6.3.14); K02154 V-type H+-transp... 36.2 0.031
ath:AT2G28520 VHA-A1; VHA-A1 (VACUOLAR PROTON ATPASE A 1); ATP... 35.0 0.057
ath:AT3G43930 hypothetical protein 34.7 0.082
xla:734990 rabgap1l, MGC130926, hhl, tbc1d18; RAB GTPase activ... 33.5 0.20
xla:379167 hypothetical protein MGC52980 32.0 0.48
tgo:TGME49_049940 hypothetical protein 30.4 1.5
mmu:27060 Tcirg1, ATP6N1C, ATP6a3, Atp6i, OC-116, OPTB1, Stv1,... 30.4 1.6
dre:560122 si:ch211-106h11.4 30.0 2.0
dre:678620 MGC136767; zgc:136767 30.0 2.3
cel:H37N21.1 hypothetical protein; K08875 nuclear receptor-bin... 29.3 3.7
hsa:9696 CROCC, KIAA0445, ROLT; ciliary rootlet coiled-coil, r... 29.3 3.7
dre:559593 aggrecan core protein-like 28.5 5.3
dre:571944 ret finger protein-like 28.5 5.5
mmu:230872 Crocc, KIAA0445; ciliary rootlet coiled-coil, rootl... 28.1 6.8
hsa:100287045 family with sequence similarity 90, member A-like 28.1 7.0
ath:AT4G11610 C2 domain-containing protein 28.1 7.1
cpv:cgd5_2180 hypothetical protein 28.1 7.2
xla:495115 tsr1; TSR1, 20S rRNA accumulation, homolog; K14799 ... 28.1 7.4
dre:100151764 btr06, zgc:172052; bloodthirsty-related gene fam... 28.1 7.6
hsa:8701 DNAH11, CILD7, DNAHBL, DNAHC11, DNHBL, DPL11, FLJ3009... 27.7 9.7
> tgo:TGME49_032830 vacuolar proton-translocating ATPase subunit,
putative (EC:3.6.3.14); K02154 V-type H+-transporting ATPase
subunit I [EC:3.6.3.14]
Length=909
Score = 179 bits (453), Expect = 2e-45, Method: Compositional matrix adjust.
Identities = 82/134 (61%), Positives = 100/134 (74%), Gaps = 0/134 (0%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDV 60
D TG +V + VFV+Y+QG++ SLL EKI KV AF+ + YEWP + EA RL L +
Sbjct 251 DSNTGNEVEKGVFVIYYQGAAHSLLREKIVKVCAAFDAKPYEWPHSAEEAATRLAGLQSL 310
Query 61 IKDKEKALAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALNCFEGHDMT 120
+ DKE+ALAAYE YFL EIS LLEV R GGSSL+EEW+MFC KEKA+YA LN F+G DMT
Sbjct 311 LDDKERALAAYEKYFLSEISLLLEVTRPGGSSLLEEWKMFCQKEKAVYATLNQFQGKDMT 370
Query 121 LRCTCWIPKKNEEK 134
LRC CWIP+ EE+
Sbjct 371 LRCDCWIPRDKEEE 384
> cpv:cgd4_1470 vacuolar proton translocating ATpase with 7 transmembrane
regions near C-terminus ; K02154 V-type H+-transporting
ATPase subunit I [EC:3.6.3.14]
Length=920
Score = 156 bits (394), Expect = 2e-38, Method: Composition-based stats.
Identities = 71/135 (52%), Positives = 94/135 (69%), Gaps = 1/135 (0%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDV 60
DPKT + V + VFV+Y QG++ S +++KI ++ +AFN Y WP ++ A R+ L+ +
Sbjct 249 DPKTSKDVQKVVFVIYFQGATTSAVYDKISRICDAFNVSIYPWPSSYEHAIQRISELNTL 308
Query 61 IKDKEKALAAYEYYFLGEISTLLE-VPREGGSSLIEEWRMFCSKEKAIYAALNCFEGHDM 119
I+DKEKAL AYE Y EI TLL+ V G+SLIEEWR+FC KEK+IYA LN FEG D+
Sbjct 309 IQDKEKALQAYEQYITLEIETLLQPVNSNNGNSLIEEWRLFCIKEKSIYATLNLFEGSDI 368
Query 120 TLRCTCWIPKKNEEK 134
TLR CW P + EEK
Sbjct 369 TLRADCWYPTEEEEK 383
> pfa:PF08_0113 vacuolar proton translocating ATPase subunit A,
putative; K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=1053
Score = 148 bits (374), Expect = 4e-36, Method: Composition-based stats.
Identities = 66/124 (53%), Positives = 90/124 (72%), Gaps = 0/124 (0%)
Query 11 SVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDVIKDKEKALAA 70
SVFVVY QGS+QS +++KI K+ +A++ + Y+WP+T+ A+ RL+ L ++I DKEKAL A
Sbjct 293 SVFVVYCQGSAQSNIYDKIMKICKAYDVKTYDWPRTYEHAKKRLKELREIINDKEKALKA 352
Query 71 YEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALNCFEGHDMTLRCTCWIPKK 130
YE YF+ EI L+ V +SLIEEW++FC KE+ IY LN FEG D+TLRC CW
Sbjct 353 YEEYFINEIFVLINVVEPNKNSLIEEWKLFCKKERHIYNNLNYFEGSDITLRCDCWYSAN 412
Query 131 NEEK 134
+EEK
Sbjct 413 DEEK 416
> tpv:TP04_0731 vacuolar ATP synthase subunit A (EC:3.6.3.14);
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=936
Score = 134 bits (338), Expect = 6e-32, Method: Compositional matrix adjust.
Identities = 65/125 (52%), Positives = 84/125 (67%), Gaps = 0/125 (0%)
Query 10 RSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDVIKDKEKALA 69
++VFV+Y Q S+ + KIKK+ F + + W +T E RL+ L DVIKDK++AL
Sbjct 259 KTVFVIYCQSSNNDATYNKIKKLCTGFQAKLFNWCKTQSELAPRLKTLEDVIKDKKRALE 318
Query 70 AYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALNCFEGHDMTLRCTCWIPK 129
AY+ YF GEI+ LLEV R GG+S+IEEW +FC KEK +Y LN FEG D+TLR CW P
Sbjct 319 AYKDYFRGEIACLLEVIRPGGNSVIEEWFLFCKKEKYLYYILNHFEGSDITLRADCWFPA 378
Query 130 KNEEK 134
EEK
Sbjct 379 DEEEK 383
> tgo:TGME49_090720 vacuolar proton-translocating ATPase subunit,
putative (EC:3.6.3.14); K02154 V-type H+-transporting ATPase
subunit I [EC:3.6.3.14]
Length=1015
Score = 131 bits (330), Expect = 4e-31, Method: Composition-based stats.
Identities = 65/133 (48%), Positives = 82/133 (61%), Gaps = 8/133 (6%)
Query 10 RSVFVVYHQG--SSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDVIKDKEKA 67
+S+FV Y+QG S S +EK+ ++ AF +CY WP +F EAE R +S ++ DKEK
Sbjct 305 KSLFVTYYQGGCSPSSAAYEKVIRLCAAFGARCYSWPGSFEEAEKRFADVSSLLADKEKT 364
Query 68 LAAYEYYFLGEISTLLEV------PREGGSSLIEEWRMFCSKEKAIYAALNCFEGHDMTL 121
L AYE YFL EIS LLE R LIE WR FC KEKA+YA LN FE D+T+
Sbjct 365 LRAYEQYFLSEISILLEPVDADCEGRRRRRPLIEAWRRFCVKEKAVYATLNFFEASDVTI 424
Query 122 RCTCWIPKKNEEK 134
R CW P ++E K
Sbjct 425 RADCWFPAQDEAK 437
> bbo:BBOV_III008300 17.m07725; V-type ATPase 116kDa subunit family
protein; K02154 V-type H+-transporting ATPase subunit
I [EC:3.6.3.14]
Length=927
Score = 123 bits (308), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 57/122 (46%), Positives = 81/122 (66%), Gaps = 0/122 (0%)
Query 12 VFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDVIKDKEKALAAY 71
VFV+Y Q +S S +K++K+ F + + W ++ RLQ L ++I+D++KAL A+
Sbjct 265 VFVIYCQSASGSSTFQKLQKLCNGFQAKTFAWSKSHSHINQRLQELEEIIRDRQKALNAF 324
Query 72 EYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALNCFEGHDMTLRCTCWIPKKN 131
+ YF EI+ LLE PR G+S+IEEW +FC KEK IY LN FEG D+TLR CW P++
Sbjct 325 KRYFREEIACLLECPRPDGNSVIEEWSLFCRKEKYIYYILNHFEGSDITLRADCWFPEEE 384
Query 132 EE 133
EE
Sbjct 385 EE 386
> dre:553691 atp6v0a1b, MGC112214, wu:fj37e01, zgc:112214; ATPase,
H+ transporting, lysosomal V0 subunit a isoform 1b; K02154
V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=839
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 39/132 (29%), Positives = 61/132 (46%), Gaps = 17/132 (12%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDV 60
DP TG QVH+SVF+++ QG L ++KK+ E F Y P+T +E ++ ++
Sbjct 208 DPTTGDQVHKSVFIIFFQGDQ---LKNRVKKICEGFRASLYPCPETPQERKEMASGVNTR 264
Query 61 IKDKEKALAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALN-CFEGHDM 119
I D + L E + + + + + W + K KAIY LN C D+
Sbjct 265 IDDLQMVLNQTEDH--------RQRVLQAAAKTMRVWFIKVRKMKAIYHTLNLC--NIDV 314
Query 120 TLRCT---CWIP 128
T +C W P
Sbjct 315 TQKCLIAEVWCP 326
> dre:324307 atp6v0a1a, atp6v0a1, wu:fc25b09, zgc:76965; ATPase,
H+ transporting, lysosomal V0 subunit a isoform 1a (EC:3.6.3.14);
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=834
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 38/132 (28%), Positives = 60/132 (45%), Gaps = 17/132 (12%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDV 60
DP TG VH+SVF+++ QG L ++KK+ E F Y P+T +E ++ ++
Sbjct 208 DPATGDNVHKSVFIIFFQGDQ---LKNRVKKICEGFRASLYPCPETPQERKEMAAGVNTR 264
Query 61 IKDKEKALAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALN-CFEGHDM 119
I D + L E + + + + + W + K KAIY LN C D+
Sbjct 265 IDDLQMVLNQTEDH--------RQRVLQAAAKTVRVWFIKVRKMKAIYHTLNLC--NIDV 314
Query 120 TLRCT---CWIP 128
T +C W P
Sbjct 315 TQKCLIAEIWCP 326
> mmu:11975 Atp6v0a1, AA959968, ATP6a1, Atp6n1, Atp6n1a, Atpv0a1,
Vpp-1, Vpp1; ATPase, H+ transporting, lysosomal V0 subunit
A1 (EC:3.6.3.14); K02154 V-type H+-transporting ATPase subunit
I [EC:3.6.3.14]
Length=838
Score = 53.1 bits (126), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 39/132 (29%), Positives = 60/132 (45%), Gaps = 17/132 (12%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDV 60
DP TG VH+SVF+++ QG L ++KK+ E F Y P+T +E ++ ++
Sbjct 208 DPVTGDYVHKSVFIIFFQGDQ---LKNRVKKICEGFRASLYPCPETPQERKEMASGVNTR 264
Query 61 IKDKEKALAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALN-CFEGHDM 119
I D + L E + + + + I W + K KAIY LN C D+
Sbjct 265 IDDLQMVLNQTEDH--------RQRVLQAAAKNIRVWFIKVRKMKAIYHTLNLC--NIDV 314
Query 120 TLRCT---CWIP 128
T +C W P
Sbjct 315 TQKCLIAEVWCP 326
> xla:379986 atp6v0a1, MGC52726; ATPase, H+ transporting, lysosomal
V0 subunit a1 (EC:3.6.3.14); K02154 V-type H+-transporting
ATPase subunit I [EC:3.6.3.14]
Length=831
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 38/132 (28%), Positives = 61/132 (46%), Gaps = 17/132 (12%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDV 60
DP TG VH+SVF+++ QG L ++KK+ E F Y P+T +E ++ ++
Sbjct 208 DPVTGDSVHKSVFIIFFQGDQ---LKNRVKKICEGFRASLYPCPETPQERKEMATGVNTR 264
Query 61 IKDKEKALAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALN-CFEGHDM 119
I+D + L E + + + + + W + K KAIY LN C D+
Sbjct 265 IEDLQMVLNQTEDH--------RQRVLQAAAKSLRVWFIKVRKMKAIYHTLNLC--NIDV 314
Query 120 TLRCT---CWIP 128
T +C W P
Sbjct 315 TQKCLIAEVWCP 326
> hsa:535 ATP6V0A1, ATP6N1, ATP6N1A, DKFZp781J1951, Stv1, VPP1,
Vph1, a1; ATPase, H+ transporting, lysosomal V0 subunit a1
(EC:3.6.3.14); K02154 V-type H+-transporting ATPase subunit
I [EC:3.6.3.14]
Length=838
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 39/132 (29%), Positives = 60/132 (45%), Gaps = 17/132 (12%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDV 60
DP TG VH+SVF+++ QG L ++KK+ E F Y P+T +E ++ ++
Sbjct 215 DPVTGDYVHKSVFIIFFQGDQ---LKNRVKKICEGFRASLYPCPETPQERKEMASGVNTR 271
Query 61 IKDKEKALAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALN-CFEGHDM 119
I D + L E + + + + I W + K KAIY LN C D+
Sbjct 272 IDDLQMVLNQTEDH--------RQRVLQAAAKNIRVWFIKVRKMKAIYHTLNLC--NIDV 321
Query 120 TLRCT---CWIP 128
T +C W P
Sbjct 322 TQKCLIAEVWCP 333
> cel:F35H10.4 vha-5; Vacuolar H ATPase family member (vha-5);
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=873
Score = 48.9 bits (115), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 36/137 (26%), Positives = 66/137 (48%), Gaps = 16/137 (11%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYE-WPQTFREAEDRLQALSD 59
DP TG++VH+SVF+++ +G + ++KV + F + ++ P+TF+E + +
Sbjct 210 DPGTGEKVHKSVFIIFLKGDR---MRSIVEKVCDGFKAKLFKNCPKTFKERQSARNDVRA 266
Query 60 VIKDKEKALA-AYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALN--CFEG 116
I+D + L E+ F +L+ +++ RM K ++ LN F+G
Sbjct 267 RIQDLQTVLGQTREHRF-----RVLQAAANNHHQWLKQVRMI----KTVFHMLNLFTFDG 317
Query 117 HDMTLRCTCWIPKKNEE 133
CWIP K+ E
Sbjct 318 IGRFFVGECWIPLKHVE 334
> cel:C26H9A.1 vha-7; Vacuolar H ATPase family member (vha-7);
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=1213
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 37/141 (26%), Positives = 63/141 (44%), Gaps = 26/141 (18%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQT-------FREAEDR 53
DP T + + + VF+V+ +G S L+ ++KV + FN Y P++ E E R
Sbjct 524 DPVTLEPLQKCVFIVFFKGESLRLI---VEKVCDGFNATQYPCPKSSKDRKMKMSETEGR 580
Query 54 LQALSDVIKDKEKALAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALNC 113
+ L+ VI + + Y L ++S E+P W +K+++A +N
Sbjct 581 MNDLTVVIDTTQ----THRYTILKDMS--FEIPI---------WLKNIQIQKSVFAVMNM 625
Query 114 FE-GHDMTLRCTCWIPKKNEE 133
F + L CWIP E+
Sbjct 626 FTVDTNGFLAGECWIPAAEED 646
> ath:AT2G21410 VHA-A2; VHA-A2 (VACUOLAR PROTON ATPASE A2); ATPase;
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=821
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 36/131 (27%), Positives = 60/131 (45%), Gaps = 15/131 (11%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDV 60
DP +G++ ++VFVV++ G KI K+ EAF Y + + + + +S
Sbjct 232 DPNSGEKAEKNVFVVFYSGERAK---SKILKICEAFGANRYPFSEDLGKQAQMMTEVSGR 288
Query 61 IKDKEKALAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALNCFEGHDMT 120
+ + + + A L + + LLE + E+W + KEKAIY LN D+T
Sbjct 289 LSELKTTIGAG----LDQRNILLETIGDK----FEQWNLKIRKEKAIYHTLNML-SLDVT 339
Query 121 LRCTC---WIP 128
+C W P
Sbjct 340 KKCLVGEGWSP 350
> ath:AT4G39080 VHA-A3; VHA-A3 (VACUOLAR PROTON ATPASE A3); ATPase;
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=821
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 36/131 (27%), Positives = 59/131 (45%), Gaps = 15/131 (11%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDV 60
DP +G++ ++VFVV++ G KI K+ EAF Y + + + +S
Sbjct 231 DPNSGEKAEKNVFVVFYSGERAK---SKILKICEAFGANRYPFSEDLGRQAQMITEVSGR 287
Query 61 IKDKEKALAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALNCFEGHDMT 120
+ + + + A LG+ + LL+ + E W + KEKAIY LN D+T
Sbjct 288 LSELKTTIDAG----LGQRNILLQTIGDK----FELWNLKVRKEKAIYHTLNML-SLDVT 338
Query 121 LRCTC---WIP 128
+C W P
Sbjct 339 KKCLVAEGWSP 349
> sce:YOR270C VPH1; Subunit a of vacuolar-ATPase V0 domain, one
of two isoforms (Vph1p and Stv1p); Vph1p is located in V-ATPase
complexes of the vacuole while Stv1p is located in V-ATPase
complexes of the Golgi and endosomes (EC:3.6.3.14); K02154
V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=840
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 36/131 (27%), Positives = 62/131 (47%), Gaps = 13/131 (9%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDV 60
D KT + H++ F+V+ G L+ ++I+K+AE+ + Y+ + E R Q L+ V
Sbjct 224 DVKTREYKHKNAFIVFSHGD---LIIKRIRKIAESLDANLYDVDSS---NEGRSQQLAKV 277
Query 61 IKDKEKALAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALN--CFEGHD 118
K+ Y L ST LE + ++ W ++EKAI+ LN ++ +
Sbjct 278 NKNLSDL-----YTVLKTTSTTLESELYAIAKELDSWFQDVTREKAIFEILNKSNYDTNR 332
Query 119 MTLRCTCWIPK 129
L WIP+
Sbjct 333 KILIAEGWIPR 343
> xla:398897 atp6v0a4, MGC68661; ATPase, H+ transporting, lysosomal
V0 subunit a4 (EC:3.6.3.14); K02154 V-type H+-transporting
ATPase subunit I [EC:3.6.3.14]
Length=846
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 38/138 (27%), Positives = 63/138 (45%), Gaps = 17/138 (12%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDV 60
DP T ++V ++VF++++QG L KIKK+ + F Y ++ E ++ ++
Sbjct 212 DPITKEEVKKNVFIIFYQGDQLKL---KIKKICDGFKATVYPCSESATERKEMAADVNTR 268
Query 61 IKDKEKALAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALN-CFEGHDM 119
I+D + E S V E SL W + K KA+Y LN C D+
Sbjct 269 IEDLNTVITQTE-------SHRQRVLLEAAQSLC-NWSIKVKKMKAVYHVLNLC--NIDV 318
Query 120 TLRCT---CWIPKKNEEK 134
T +C W P ++E+
Sbjct 319 TQQCVIAEIWCPVSDKER 336
> hsa:50617 ATP6V0A4, A4, ATP6N1B, ATP6N2, MGC130016, MGC130017,
RDRTA2, RTA1C, RTADR, STV1, VPH1, VPP2; ATPase, H+ transporting,
lysosomal V0 subunit a4 (EC:3.6.3.14); K02154 V-type
H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=840
Score = 44.3 bits (103), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 33/138 (23%), Positives = 65/138 (47%), Gaps = 17/138 (12%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDV 60
DP T +++ +++F++++QG L +KIKK+ + F Y P+ E + L++++
Sbjct 210 DPVTKEEIQKNIFIIFYQGEQ---LRQKIKKICDGFRATVYPCPEPAVERREMLESVNVR 266
Query 61 IKDKEKALAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALN-CFEGHDM 119
++D + E S + +E ++ W + K KA+Y LN C D+
Sbjct 267 LEDLITVITQTE-------SHRQRLLQEAAANW-HSWLIKVQKMKAVYHILNMC--NIDV 316
Query 120 TLRCT---CWIPKKNEEK 134
T +C W P + +
Sbjct 317 TQQCVIAEIWFPVADATR 334
> cel:ZK637.8 unc-32; UNCoordinated family member (unc-32); K02154
V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=899
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 36/131 (27%), Positives = 53/131 (40%), Gaps = 15/131 (11%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDV 60
D TG V++ VF+++ QG L K+KK+ E F Y P T +E + +
Sbjct 244 DTVTGDPVNKCVFIIFFQGDH---LKTKVKKICEGFRATLYPCPDTPQERREMSIGVMTR 300
Query 61 IKDKEKALAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALNCFEGHDMT 120
I+D + LG+ S + W K K+IY LN F D+T
Sbjct 301 IEDLKTV--------LGQTQDHRHRVLVAASKNVRMWLTKVRKIKSIYHTLNLFN-IDVT 351
Query 121 LRCT---CWIP 128
+C W P
Sbjct 352 QKCLIAEVWCP 362
> dre:100003139 tcirg1, si:dkey-9i23.9; T-cell, immune regulator
1, ATPase, H+ transporting, lysosomal V0 subunit A3; K02154
V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=822
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 30/113 (26%), Positives = 52/113 (46%), Gaps = 13/113 (11%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDV 60
+P TG+ + +VFV+ G + +K+KK+ + F + +P+ E E LQ L
Sbjct 207 EPDTGETIQWTVFVISFWGEQ---IGQKVKKICDCFRTHTFVYPEGLEEREGILQGLESR 263
Query 61 IKDKEKALAAYEYYFLGEIST-LLEVPREGGSSLIEEWRMFCSKEKAIYAALN 112
I D L+ E Y +S + ++P +W++ K KA+ LN
Sbjct 264 IVDIRTVLSQTEQYMQQLLSRCVCQMP---------QWKIRVQKCKAVQMVLN 307
> dre:561117 si:ch211-106a19.2 (EC:3.6.3.6); K02154 V-type H+-transporting
ATPase subunit I [EC:3.6.3.14]
Length=849
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 34/133 (25%), Positives = 58/133 (43%), Gaps = 13/133 (9%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDV 60
DP +G+ VF++ + G + +K+KK+ + ++ Y +P + E D ++ L
Sbjct 216 DPDSGEPTRSVVFLISYWGEQ---IGQKVKKICDCYHCHLYPYPNSNEERTDVVEGLRTR 272
Query 61 IKDKEKALAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALNC--FEGHD 118
I+D L E Y L +V + S + W + K KAIY LN F+ +
Sbjct 273 IQDLHTVLHRTEDY-------LRQVLIKASES-VYIWVIQVKKMKAIYHILNLCSFDVTN 324
Query 119 MTLRCTCWIPKKN 131
L W P +
Sbjct 325 KCLIAEVWCPVND 337
> dre:100332691 atp6v0a1; ATPase, H+ transporting, lysosomal V0
subunit a1
Length=803
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 32/105 (30%), Positives = 48/105 (45%), Gaps = 11/105 (10%)
Query 8 VHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDVIKDKEKA 67
V + F+++ QG + EKI+KV E F Y P+T E ++ ++ ++D
Sbjct 227 VKKDAFIIFVQGDH---VREKIRKVCEGFRASLYSCPKTLYERKEMSNSIMTRMEDLRLV 283
Query 68 LAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALN 112
L E Y G +S E +E GS + K KAIY LN
Sbjct 284 LRRTEEYRAGVLSRAAEHVQEWGSKV--------KKMKAIYYTLN 320
> mmu:140494 Atp6v0a4, Atp6n1b, a4; ATPase, H+ transporting, lysosomal
V0 subunit A4 (EC:3.6.3.14); K02154 V-type H+-transporting
ATPase subunit I [EC:3.6.3.14]
Length=833
Score = 41.6 bits (96), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 33/132 (25%), Positives = 61/132 (46%), Gaps = 17/132 (12%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDV 60
DP T +++ +++F++++QG L KIKK+ + F Y P+ E + L +++
Sbjct 210 DPVTKEEIKKNIFIIFYQGEQLRL---KIKKICDGFRATIYPCPEHAAERREMLTSVNVR 266
Query 61 IKDKEKALAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALN-CFEGHDM 119
++D + E S + +E ++ W + K KA+Y LN C D+
Sbjct 267 LEDLITVITQTE-------SHRQRLLQEAAANW-HSWVIKVQKMKAVYHVLNMC--NIDV 316
Query 120 TLRCT---CWIP 128
T +C W P
Sbjct 317 TQQCIIAEIWFP 328
> dre:100147923 T-cell immune regulator 1-like; K02154 V-type
H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=793
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 28/111 (25%), Positives = 51/111 (45%), Gaps = 11/111 (9%)
Query 2 PKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDVI 61
P T +Q+ +VF++ G + +K+KK+ + F+ Q + +P+ E E+ L L I
Sbjct 204 PHTDEQLQWTVFLISFWGDQ---IGQKVKKICDCFHTQTFPYPENQAEREETLNGLRGRI 260
Query 62 KDKEKALAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALN 112
+D + + E Y + L + + EW + K KA+ LN
Sbjct 261 EDIKSVMGETEQYMQQLLVRAL--------ARLPEWVVQVQKCKAVQTVLN 303
> dre:406342 tcirg1, wu:fb44e06, wu:fi46h07; zgc:55891; K02154
V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=822
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 28/111 (25%), Positives = 51/111 (45%), Gaps = 11/111 (9%)
Query 2 PKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDVI 61
P T +Q+ +VF++ G + +K+KK+ + F+ Q + +P+ E E+ L L I
Sbjct 204 PHTDEQLQWTVFLISFWGDQ---IGQKVKKICDCFHTQTFPYPENQAEREETLNGLRGRI 260
Query 62 KDKEKALAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALN 112
+D + + E Y + L + + EW + K KA+ LN
Sbjct 261 EDIKSVMGETEQYMQQLLVRAL--------ARLPEWVVQVQKCKAVQTVLN 303
> cel:VW02B12L.1 vha-6; Vacuolar H ATPase family member (vha-6);
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=865
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 29/125 (23%), Positives = 53/125 (42%), Gaps = 15/125 (12%)
Query 10 RSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDVIKDKEKALA 69
+ VF+++ G L K+KK+ + F +CY P+ E L + D + +
Sbjct 228 KCVFILFFSGEQ---LRAKVKKICDGFQAKCYTVPENPAERTKLLLNIKVQTTDMKAVIE 284
Query 70 AYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALNCFEGHDMTLRCT---CW 126
Y I ++ + +W + K K+I+ LN F D+T +C CW
Sbjct 285 KTLDYRSKCI--------HAAATNLRKWGIMLLKLKSIFHTLNMFSV-DVTQKCLIAECW 335
Query 127 IPKKN 131
+P+ +
Sbjct 336 VPEAD 340
> mmu:21871 Atp6v0a2, 8430408C20Rik, AI385560, ATP6a2, AW489264,
Atp6n1d, Atp6n2, C76904, MGC124341, MGC124342, Stv1, TJ6M,
TJ6s, Tj6; ATPase, H+ transporting, lysosomal V0 subunit A2
(EC:3.6.3.6); K02154 V-type H+-transporting ATPase subunit
I [EC:3.6.3.14]
Length=856
Score = 37.0 bits (84), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 35/131 (26%), Positives = 58/131 (44%), Gaps = 13/131 (9%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDV 60
DP+TG+ + VF++ G + K+KK+ + ++ Y +P T E + + L+
Sbjct 213 DPETGEVIKWYVFLISFWGEQ---IGHKVKKICDCYHCHIYPYPNTAEERREIQEGLNTR 269
Query 61 IKDKEKALAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALNC--FEGHD 118
I+D L E Y + +L E S + + R K KAIY LN F+ +
Sbjct 270 IQDLYTVLHKTEDY----LRQVLCKAAESVCSRVVQVR----KMKAIYHMLNMCSFDVTN 321
Query 119 MTLRCTCWIPK 129
L W P+
Sbjct 322 KCLIAEVWCPE 332
> dre:561469 ATPase, H+ transporting, lysosomal V0 subunit a1-like;
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=839
Score = 36.2 bits (82), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 34/133 (25%), Positives = 55/133 (41%), Gaps = 13/133 (9%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDV 60
D TG+ VF++ G + +K++K+ + ++ Y P+T E D + +L
Sbjct 213 DLDTGEIRKNVVFLISFWGDQ---IGQKVQKICDCYHCHIYPHPETDEERADVMDSLRTR 269
Query 61 IKDKEKALAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALNC--FEGHD 118
I+D L E Y + +L E S W + K KAIY LN F+ +
Sbjct 270 IQDLHNVLHRTEDY----LKQVLHKASESAQS----WVLQVKKMKAIYHILNLCSFDVTN 321
Query 119 MTLRCTCWIPKKN 131
L W P +
Sbjct 322 KCLIAEVWCPVSD 334
> hsa:23545 ATP6V0A2, A2, ARCL, ATP6A2, ATP6N1D, J6B7, RTF, STV1,
TJ6, TJ6M, TJ6S, VPH1, WSS; ATPase, H+ transporting, lysosomal
V0 subunit a2 (EC:3.6.3.6); K02154 V-type H+-transporting
ATPase subunit I [EC:3.6.3.14]
Length=856
Score = 36.2 bits (82), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 34/133 (25%), Positives = 57/133 (42%), Gaps = 13/133 (9%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDV 60
DP+TG+ + VF++ G + K+KK+ + ++ Y +P T E + + L+
Sbjct 213 DPETGEVIKWYVFLISFWGEQ---IGHKVKKICDCYHCHVYPYPNTAEERREIQEGLNTR 269
Query 61 IKDKEKALAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALNC--FEGHD 118
I+D L E Y L + S +I+ K KAIY LN F+ +
Sbjct 270 IQDLYTVLHKTEDYLR---QVLCKAAESVYSRVIQ-----VKKMKAIYHMLNMCSFDVTN 321
Query 119 MTLRCTCWIPKKN 131
L W P+ +
Sbjct 322 KCLIAEVWCPEAD 334
> sce:YMR054W STV1; Stv1p (EC:3.6.3.14); K02154 V-type H+-transporting
ATPase subunit I [EC:3.6.3.14]
Length=890
Score = 36.2 bits (82), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 27/123 (21%), Positives = 50/123 (40%), Gaps = 14/123 (11%)
Query 6 QQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDVIKDKE 65
++V + F+++ G + L +K+K+V ++ NG+ E D L+ I D +
Sbjct 280 EKVEKDCFIIFTHGET---LLKKVKRVIDSLNGKIVSLNTRSSELVD---TLNRQIDDLQ 333
Query 66 KALAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALNCFEGHDMTLRCTC 125
+ L E E+ + + W +EK +Y LN F+ L
Sbjct 334 RILDTTEQTLHTELLVI--------HDQLPVWSAMTKREKYVYTTLNKFQQESQGLIAEG 385
Query 126 WIP 128
W+P
Sbjct 386 WVP 388
> ath:AT2G28520 VHA-A1; VHA-A1 (VACUOLAR PROTON ATPASE A 1); ATPase;
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=817
Score = 35.0 bits (79), Expect = 0.057, Method: Compositional matrix adjust.
Identities = 37/131 (28%), Positives = 55/131 (41%), Gaps = 15/131 (11%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDV 60
DP T + V + VFVV+ G KI K+ EAF CY P+ + + +
Sbjct 230 DPSTSEMVEKVVFVVFFSGEQA---RTKILKICEAFGANCYPVPEDTTKQRQLTREVLSR 286
Query 61 IKDKEKALAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALNCFEGHDMT 120
+ D E L A + ++++ G SL W +EKA+Y LN D+T
Sbjct 287 LSDLEATLDAGTRHRNNALNSV-------GYSL-TNWITTVRREKAVYDTLNMLN-FDVT 337
Query 121 LRCTC---WIP 128
+C W P
Sbjct 338 KKCLVGEGWCP 348
> ath:AT3G43930 hypothetical protein
Length=444
Score = 34.7 bits (78), Expect = 0.082, Method: Compositional matrix adjust.
Identities = 22/83 (26%), Positives = 39/83 (46%), Gaps = 7/83 (8%)
Query 59 DVIKDKEKALAAYEYYFLGEIS-----TLLEVPREGGSSLIEEWRMFCSKEKAI--YAAL 111
+V K+ AL+ Y + +IS + +V EGG+ L+ +W + C+ + ++
Sbjct 176 EVTGSKDLALSGYSVFIDADISEEVRRQVSQVAVEGGAKLMTQWFIGCNASHVVCEGGSV 235
Query 112 NCFEGHDMTLRCTCWIPKKNEEK 134
+ GH L W+ K EEK
Sbjct 236 LRYLGHSSNLVTPLWLQKTLEEK 258
> xla:734990 rabgap1l, MGC130926, hhl, tbc1d18; RAB GTPase activating
protein 1-like
Length=1055
Score = 33.5 bits (75), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 30/128 (23%), Positives = 51/128 (39%), Gaps = 20/128 (15%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDV 60
DPK+ + +S V+ H +S+ +F+ CY + ++AL
Sbjct 102 DPKSTELRVQSTSVLSHPPEEESV----------SFHKLCYLGCMKVSAPRNEIEAL--- 148
Query 61 IKDKEKALAAYEYYFLGEISTLLEVPR--EGGSSLIEEWRMFCSKEKAIYAALNCFEGHD 118
+A+A+ + + I+ L VP EG +I++ IY L C G D
Sbjct 149 -----RAMASMKAQCISPITVTLYVPNIPEGSVRIIDQTNKSEIASFPIYKVLFCVRGRD 203
Query 119 MTLRCTCW 126
T C C+
Sbjct 204 GTSECDCF 211
> xla:379167 hypothetical protein MGC52980
Length=1052
Score = 32.0 bits (71), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 29/128 (22%), Positives = 51/128 (39%), Gaps = 20/128 (15%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDV 60
DPK+ + +S+ + H +S+ +F+ CY + ++AL
Sbjct 102 DPKSTELPVQSIGALSHPPEEESV----------SFHKLCYLGCMKVSAPRNEIEAL--- 148
Query 61 IKDKEKALAAYEYYFLGEISTLLEVPR--EGGSSLIEEWRMFCSKEKAIYAALNCFEGHD 118
+A+A+ + + I+ L VP EG +I++ IY L C G D
Sbjct 149 -----RAMASKKAQCVAPITVTLYVPNIPEGSVRIIDQTNKSEVASFPIYKVLFCVRGRD 203
Query 119 MTLRCTCW 126
T C C+
Sbjct 204 GTSECDCF 211
> tgo:TGME49_049940 hypothetical protein
Length=118
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 21/42 (50%), Gaps = 0/42 (0%)
Query 91 SSLIEEWRMFCSKEKAIYAALNCFEGHDMTLRCTCWIPKKNE 132
+SL+EE F S E+A+ +N G +TL C P E
Sbjct 67 TSLLEEDLQFTSAEEALSLTVNTLFGRGLTLEKRCLTPTTQE 108
> mmu:27060 Tcirg1, ATP6N1C, ATP6a3, Atp6i, OC-116, OPTB1, Stv1,
TIRC7, Vph1, oc; T-cell, immune regulator 1, ATPase, H+ transporting,
lysosomal V0 protein A3 (EC:3.6.3.6); K02154 V-type
H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=834
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 29/112 (25%), Positives = 49/112 (43%), Gaps = 11/112 (9%)
Query 1 DPKTGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDV 60
DP TG+ FV+ + G + +KI+K+ + F+ C+ +P +E E R + L
Sbjct 208 DPVTGEPATWMTFVISYWGEQ---IGQKIRKITDCFH--CHVFPYLEQE-EARFRTLQ-- 259
Query 61 IKDKEKALAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFCSKEKAIYAALN 112
+ + + LGE L L+ W++ K KA+Y LN
Sbjct 260 ---QLQQQSQELQEVLGETDRFLSQVLGRVQQLLPPWQVQIHKMKAVYLTLN 308
> dre:560122 si:ch211-106h11.4
Length=455
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 45 QTFREAEDRLQALSDVIKDKEKALAAYEYYFLGEISTLLE 84
+ E + ALSD I+D E+ L A + FL E L+E
Sbjct 209 EKLEEINTHISALSDSIRDTEEMLKANDVCFLKEFPGLME 248
> dre:678620 MGC136767; zgc:136767
Length=356
Score = 30.0 bits (66), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 45 QTFREAEDRLQALSDVIKDKEKALAAYEYYFLGEISTLLE 84
+T E E ++ ++D IKD E+ L A + L E L+E
Sbjct 242 KTVEEIESQISTITDAIKDTEEMLKASDDCLLKEADVLME 281
> cel:H37N21.1 hypothetical protein; K08875 nuclear receptor-binding
protein
Length=649
Score = 29.3 bits (64), Expect = 3.7, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 32/62 (51%), Gaps = 4/62 (6%)
Query 58 SDVIKDKEKALAAYEYYFLGEISTLLEVPREGGSSL-IEEWRMFCSKEKAIYAALNCFEG 116
+D +K + + EY G +S L+ R+ GSSL I+ W+ + ++ I +ALN
Sbjct 114 TDSKSEKPRIIFITEYMSSGSMSAFLQRTRKAGSSLSIKAWKKWTTQ---ILSALNYLHS 170
Query 117 HD 118
D
Sbjct 171 SD 172
> hsa:9696 CROCC, KIAA0445, ROLT; ciliary rootlet coiled-coil,
rootletin
Length=2017
Score = 29.3 bits (64), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 26/82 (31%), Positives = 43/82 (52%), Gaps = 4/82 (4%)
Query 23 SLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDVIKDKEKALAAYEYYFLGEISTL 82
S L E++ +++ +G+ E Q REA+ +++AL ++KE AL A E+ L L
Sbjct 830 SQLQEQLAQLSRQLSGREQELEQARREAQRQVEALERAAREKE-AL-AKEHAGLA--VQL 885
Query 83 LEVPREGGSSLIEEWRMFCSKE 104
+ REG + E R+ KE
Sbjct 886 VAAEREGRTLSEEATRLRLEKE 907
> dre:559593 aggrecan core protein-like
Length=1350
Score = 28.5 bits (62), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 22/45 (48%), Gaps = 0/45 (0%)
Query 28 KIKKVAEAFNGQCYEWPQTFREAEDRLQALSDVIKDKEKALAAYE 72
KIK V + + + T+ EA+D Q L VI E AAYE
Sbjct 432 KIKGVVFHYRAESGRYAYTYEEAQDACQKLGAVIATPELLQAAYE 476
> dre:571944 ret finger protein-like
Length=564
Score = 28.5 bits (62), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 26/48 (54%), Gaps = 0/48 (0%)
Query 22 QSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDVIKDKEKALA 69
Q ++ +++KKV E + RE ED +Q SD+I+ +KA A
Sbjct 227 QQMIQDRLKKVDEIKHSIELNKSGAQREVEDSMQVFSDLIRALQKAQA 274
> mmu:230872 Crocc, KIAA0445; ciliary rootlet coiled-coil, rootletin
Length=1845
Score = 28.1 bits (61), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 25/92 (27%), Positives = 45/92 (48%), Gaps = 5/92 (5%)
Query 20 SSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDVIKDKEKALAAYEYYFLGEI 79
S +S L E++ +++ +G+ E Q RE++ +++AL ++KE A G
Sbjct 664 SERSHLQEQLAQLSRQLSGRDQELEQALRESQRQVEALERAAREKE----AMAKERAGLA 719
Query 80 STLLEVPREGGSSLIEEWRMFCSKEKAIYAAL 111
L REG + E R+ KE A+ ++L
Sbjct 720 VKLAAAEREGRTLSEEAIRLRLEKE-ALESSL 750
> hsa:100287045 family with sequence similarity 90, member A-like
Length=464
Score = 28.1 bits (61), Expect = 7.0, Method: Composition-based stats.
Identities = 18/56 (32%), Positives = 26/56 (46%), Gaps = 3/56 (5%)
Query 15 VYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDVIKDKEKALAA 70
V HQG L+ +K + G C E PQ + LQA+S +DK A+ +
Sbjct 215 VRHQGPEPLLV---VKPTHSSPEGGCREVPQAASKTHGLLQAISPQAQDKRPAVTS 267
> ath:AT4G11610 C2 domain-containing protein
Length=1011
Score = 28.1 bits (61), Expect = 7.1, Method: Composition-based stats.
Identities = 21/59 (35%), Positives = 32/59 (54%), Gaps = 5/59 (8%)
Query 42 EWPQTFREAEDRLQA--LSDVIKDKEKALAAYEYYFLGEISTL-LEVPREGGSSLIEEW 97
EW Q F A++R+QA L V+KDK+ Y + +I+ + L VP + S L +W
Sbjct 324 EWNQVFAFAKERMQASVLEVVVKDKDLLKDDYVGFVRFDINDVPLRVPPD--SPLAPQW 380
> cpv:cgd5_2180 hypothetical protein
Length=1610
Score = 28.1 bits (61), Expect = 7.2, Method: Composition-based stats.
Identities = 24/93 (25%), Positives = 38/93 (40%), Gaps = 8/93 (8%)
Query 4 TGQQVHRSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQA------- 56
TG +V S L EK+K+ E + Y ++ + A+D L+A
Sbjct 101 TGSDAQETVRRYRDNYSQLRQLFEKLKEQNEILQDENYRLEESLKSAQDALKANRTMDSK 160
Query 57 -LSDVIKDKEKALAAYEYYFLGEISTLLEVPRE 88
LS + +K L A +Y + LL+ P E
Sbjct 161 NLSSNLNYSQKCLDACVHYISYTLKYLLDNPEE 193
> xla:495115 tsr1; TSR1, 20S rRNA accumulation, homolog; K14799
pre-rRNA-processing protein TSR1
Length=815
Score = 28.1 bits (61), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 39/75 (52%), Gaps = 6/75 (8%)
Query 1 DPKTGQQVHRSVFVVYH-QGSSQSLLHEKIKKVAEAFNGQCYEWP--QTFREAEDRLQAL 57
+P TG ++ + + V+ S+Q L ++ V + G+ WP + +EAED L+
Sbjct 336 EPATGSEMEQDIKVLMKADPSAQESLQCEV--VPDPMEGE-QTWPTEEELKEAEDALKGT 392
Query 58 SDVIKDKEKALAAYE 72
S V+K K +AY+
Sbjct 393 SKVVKKVPKGTSAYQ 407
> dre:100151764 btr06, zgc:172052; bloodthirsty-related gene family,
member 6
Length=556
Score = 28.1 bits (61), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 14/60 (23%), Positives = 30/60 (50%), Gaps = 0/60 (0%)
Query 10 RSVFVVYHQGSSQSLLHEKIKKVAEAFNGQCYEWPQTFREAEDRLQALSDVIKDKEKALA 69
+ + ++ Q Q ++ +++KK+ + + T RE RLQ SD+++ E+ A
Sbjct 208 KKIQMIKTQTDVQQMIQDRVKKIQDIEHSADLRKKSTEREKASRLQLFSDLMRSIERCQA 267
> hsa:8701 DNAH11, CILD7, DNAHBL, DNAHC11, DNHBL, DPL11, FLJ30095,
FLJ37699; dynein, axonemal, heavy chain 11
Length=4523
Score = 27.7 bits (60), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 18/73 (24%), Positives = 33/73 (45%), Gaps = 6/73 (8%)
Query 42 EWPQTFREAEDRLQALSDVIKDKEKALAAYEYYFLGEISTLLEVPREGGSSLIEEWRMFC 101
+W Q + E+ED Q L + ++ A + + G ++ E+PR+ L+ F
Sbjct 76 KWSQ-YLESEDNRQVLGEFLESTSPACLVFSFAASGRLAASQEIPRDANHKLV-----FI 129
Query 102 SKEKAIYAALNCF 114
SK+ +N F
Sbjct 130 SKKITESIGVNDF 142
Lambda K H
0.318 0.133 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2231140792
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40