bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2997_orf2
Length=167
Score E
Sequences producing significant alignments: (Bits) Value
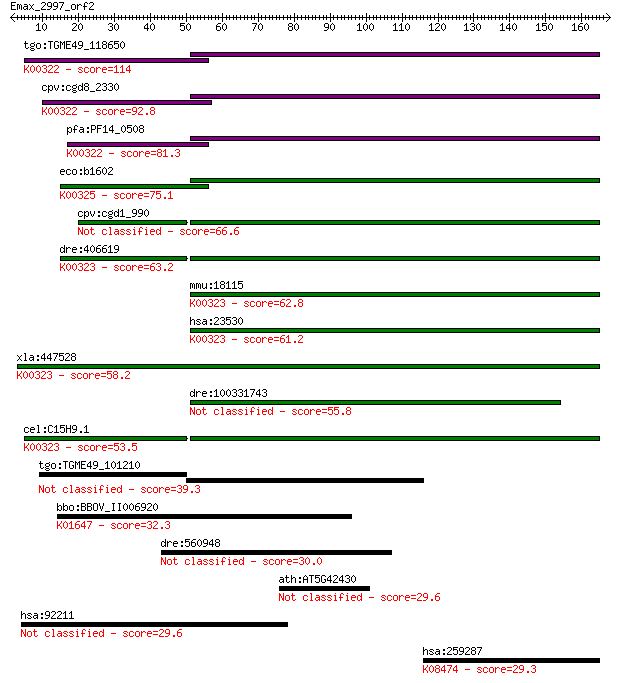
tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00... 114 1e-25
cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alp... 92.8 5e-19
pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative (... 81.3 1e-15
eco:b1602 pntB, ECK1597, JW1594; pyridine nucleotide transhydr... 75.1 9e-14
cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alph... 66.6 4e-11
dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamid... 63.2 4e-10
mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide... 62.8 5e-10
hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide t... 61.2 2e-09
xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydroge... 58.2 1e-08
dre:100331743 Nicotinamide Nucleotide Transhydrogenase family ... 55.8 6e-08
cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase fa... 53.5 3e-07
tgo:TGME49_101210 NAD(P) transhydrogenase, alpha subunit, puta... 39.3 0.006
bbo:BBOV_II006920 18.m06571; citrate synthase (EC:2.3.3.1); K0... 32.3 0.67
dre:560948 fi40d06, wu:fi40d06; zgc:174160 30.0 4.0
ath:AT5G42430 F-box family protein 29.6 5.0
hsa:92211 CDHR1, CORD15, DKFZp434A132, KIAA1775, PCDH21, PRCAD... 29.6 5.2
hsa:259287 TAS2R41, MGC119928, MGC119929, T2R41, T2R59; taste ... 29.3 7.1
> tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00322
NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1013
Score = 114 bits (286), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 59/114 (51%), Positives = 84/114 (73%), Gaps = 1/114 (0%)
Query 51 GLAAVMVGFANFHSPAGVERASSLLRLLEVYAGVFVAGITFTGSVVAAAKLHGSMESRSL 110
G AAV+VG ++FH+ G E ++ R +E G FVA +TFTGS+VAAAKLH M+ +SL
Sbjct 92 GFAAVLVGISSFHAGLG-ESGTNATRAVETAVGEFVAAVTFTGSLVAAAKLHELMDPKSL 150
Query 111 RVPGRHALNSATIAAIGVLGALFCVSSGHLTRMLCLYVNAGLSMWLGFHLVAAI 164
++PGRHA+N+ T+AA+ V+G FC ++ T+++CLY LS+WLGFHLVA+I
Sbjct 151 KIPGRHAINAMTVAAVAVVGITFCATADTTTKIICLYTLITLSLWLGFHLVASI 204
Score = 57.8 bits (138), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 28/51 (54%), Positives = 38/51 (74%), Gaps = 2/51 (3%)
Query 5 AEGVGAAAPQGAVQQTTADEVADELLAARKVLIVPGYGMAVARCQGGLAAV 55
AEGV +G V++ + D VA+E+L A+KVLIVPGYGMAV+RCQ +A +
Sbjct 275 AEGVPGV--EGVVREISPDSVAEEVLLAKKVLIVPGYGMAVSRCQSDVADI 323
> cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, possible signal peptide
plus 12 transmembrane regions ; K00322 NAD(P) transhydrogenase
[EC:1.6.1.1]
Length=1143
Score = 92.8 bits (229), Expect = 5e-19, Method: Composition-based stats.
Identities = 49/115 (42%), Positives = 74/115 (64%), Gaps = 1/115 (0%)
Query 51 GLAAVMVGFANFHSP-AGVERASSLLRLLEVYAGVFVAGITFTGSVVAAAKLHGSMESRS 109
GLAA+++ FAN +P E S + +E++ G ++ ITFTGS+VAA KLH S +
Sbjct 181 GLAAMLISFANLWTPFQSSEEEFSAVHAIEMFIGEAISAITFTGSIVAAGKLHEIFPSGA 240
Query 110 LRVPGRHALNSATIAAIGVLGALFCVSSGHLTRMLCLYVNAGLSMWLGFHLVAAI 164
L++PGRH LN+ +A + LG++F + + + R +Y N+ LSM LG HLVA+I
Sbjct 241 LKLPGRHFLNALMVAGLIALGSVFIIITDYANRTYLMYGNSLLSMLLGIHLVASI 295
Score = 50.1 bits (118), Expect = 3e-06, Method: Composition-based stats.
Identities = 25/47 (53%), Positives = 35/47 (74%), Gaps = 0/47 (0%)
Query 10 AAAPQGAVQQTTADEVADELLAARKVLIVPGYGMAVARCQGGLAAVM 56
A Q V +T A +VA +LL+A+KVLIVPGYGMAV+R Q +A+++
Sbjct 372 ALGDQSDVNKTNAMKVARDLLSAKKVLIVPGYGMAVSRSQQDVASIV 418
> pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative
(EC:1.6.1.2); K00322 NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1176
Score = 81.3 bits (199), Expect = 1e-15, Method: Composition-based stats.
Identities = 46/114 (40%), Positives = 70/114 (61%), Gaps = 0/114 (0%)
Query 51 GLAAVMVGFANFHSPAGVERASSLLRLLEVYAGVFVAGITFTGSVVAAAKLHGSMESRSL 110
GLAA+ VGF+ +HS + S + LLE+Y G F+A I F GS+VAA KL G ++S+SL
Sbjct 186 GLAALFVGFSKYHSESFENYEISTIHLLELYVGTFIASIAFIGSLVAAGKLSGILDSKSL 245
Query 111 RVPGRHALNSATIAAIGVLGALFCVSSGHLTRMLCLYVNAGLSMWLGFHLVAAI 164
++ + +N I I +LG F + +CLY++ + +LGFHL+A+I
Sbjct 246 KLQIKKIINILCIVLIIILGYYFVTLKLLYLKSICLYISLIIDSFLGFHLIASI 299
Score = 42.0 bits (97), Expect = 9e-04, Method: Composition-based stats.
Identities = 17/39 (43%), Positives = 27/39 (69%), Gaps = 0/39 (0%)
Query 17 VQQTTADEVADELLAARKVLIVPGYGMAVARCQGGLAAV 55
+ TT VA+ L+ A+ ++IVPGYG A+++CQ LA +
Sbjct 389 INSTTNKYVAENLINAKNIIIVPGYGTALSKCQRELAEI 427
> eco:b1602 pntB, ECK1597, JW1594; pyridine nucleotide transhydrogenase,
beta subunit (EC:1.6.1.2); K00325 NAD(P) transhydrogenase
subunit beta [EC:1.6.1.2]
Length=462
Score = 75.1 bits (183), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 44/115 (38%), Positives = 69/115 (60%), Gaps = 1/115 (0%)
Query 51 GLAAVMVGFANF-HSPAGVERASSLLRLLEVYAGVFVAGITFTGSVVAAAKLHGSMESRS 109
GLAAV+VGF ++ H AG+ + L EV+ G+F+ +TFTGSVVA KL G + S+
Sbjct 95 GLAAVLVGFNSYLHHDAGMAPILVNIHLTEVFLGIFIGAVTFTGSVVAFGKLCGKISSKP 154
Query 110 LRVPGRHALNSATIAAIGVLGALFCVSSGHLTRMLCLYVNAGLSMWLGFHLVAAI 164
L +P RH +N A + +L +F + ++L L + +++ G+HLVA+I
Sbjct 155 LMLPNRHKMNLAALVVSFLLLIVFVRTDSVGLQVLALLIMTAIALVFGWHLVASI 209
Score = 37.4 bits (85), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 18/41 (43%), Positives = 27/41 (65%), Gaps = 0/41 (0%)
Query 15 GAVQQTTADEVADELLAARKVLIVPGYGMAVARCQGGLAAV 55
G ++ TA+E A+ L + V+I PGYGMAVA+ Q +A +
Sbjct 289 GEHREITAEETAELLKNSHSVIITPGYGMAVAQAQYPVAEI 329
> cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, 12 transmembrane domain
(EC:1.6.1.2)
Length=1147
Score = 66.6 bits (161), Expect = 4e-11, Method: Composition-based stats.
Identities = 38/114 (33%), Positives = 65/114 (57%), Gaps = 0/114 (0%)
Query 51 GLAAVMVGFANFHSPAGVERASSLLRLLEVYAGVFVAGITFTGSVVAAAKLHGSMESRSL 110
GLAA+ V ++ F+S G++ +LR +E++ G + ITF GSV+AA KL + S+S+
Sbjct 149 GLAALFVSYSYFYSSIGMKEGIYVLRRMELFVGGVMGMITFVGSVIAALKLDDIIPSKSV 208
Query 111 RVPGRHALNSATIAAIGVLGALFCVSSGHLTRMLCLYVNAGLSMWLGFHLVAAI 164
++P ++ S + +I + G FCVS+ L L+ LS G ++ +I
Sbjct 209 KIPLKNVSLSILLFSICMTGLSFCVSNNELVITTNLHCGMILSALFGIFMIISI 262
Score = 41.2 bits (95), Expect = 0.002, Method: Composition-based stats.
Identities = 17/30 (56%), Positives = 24/30 (80%), Gaps = 0/30 (0%)
Query 20 TTADEVADELLAARKVLIVPGYGMAVARCQ 49
T++ E A+ L + K+LIVPGYGMAV++CQ
Sbjct 345 TSSKETAEILAESSKILIVPGYGMAVSKCQ 374
> dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P)
transhydrogenase [EC:1.6.1.2]
Length=1079
Score = 63.2 bits (152), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 44/118 (37%), Positives = 68/118 (57%), Gaps = 6/118 (5%)
Query 51 GLAAVMVGFANF---HSPAGVERASSLLRLLEVYAGVFVAGITFTGSVVAAAKLHGSMES 107
GLAAV+ A + + + A++L +++ Y G ++ G+TF+GS+VA KL G + S
Sbjct 707 GLAAVLTCVAEYMVEYPHFATDPAANLTKIV-AYLGTYIGGVTFSGSLVAYGKLQGLLNS 765
Query 108 RSLRVPGRHALNSATIAAIGVLGAL-FCVSSGHLTRMLCLYVNAGLSMWLGFHLVAAI 164
L +PGRHALN AT+ A V G + + + + T + CL + LS +G L AAI
Sbjct 766 APLMLPGRHALN-ATLMAASVGGMIPYMLDPSYTTGITCLGSVSALSAVMGLTLTAAI 822
Score = 31.6 bits (70), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 12/35 (34%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 15 GAVQQTTADEVADELLAARKVLIVPGYGMAVARCQ 49
G + D+ D + A ++IVPGYG+ A+ Q
Sbjct 903 GTHTEVNVDQTVDLIKEAHNIIIVPGYGLCAAKAQ 937
> mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 62.8 bits (151), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 45/117 (38%), Positives = 63/117 (53%), Gaps = 4/117 (3%)
Query 51 GLAAVMVGFANF--HSPAGVERASSLLRLLEVYAGVFVAGITFTGSVVAAAKLHGSMESR 108
GLAAV+ A + P A+S + Y G ++ G+TF+GS+VA KL G ++S
Sbjct 711 GLAAVLTCMAEYIVEYPHFAMDATSNFTKIVAYLGTYIGGVTFSGSLVAYGKLQGILKSA 770
Query 109 SLRVPGRHALNSATIAAIGVLGAL-FCVSSGHLTRMLCLYVNAGLSMWLGFHLVAAI 164
L +PGRHALN+ +AA V G + F T + CL + LS +G L AAI
Sbjct 771 PLLLPGRHALNAGLLAA-SVGGIIPFMADPSFTTGITCLGSVSALSTLMGVTLTAAI 826
> hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide
transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 61.2 bits (147), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 44/117 (37%), Positives = 64/117 (54%), Gaps = 4/117 (3%)
Query 51 GLAAVMVGFANF--HSPAGVERASSLLRLLEVYAGVFVAGITFTGSVVAAAKLHGSMESR 108
GLAAV+ A + P A++ L + Y G ++ G+TF+GS++A KL G ++S
Sbjct 711 GLAAVLTCIAEYIIEYPHFATDAAANLTKIVAYLGTYIGGVTFSGSLIAYGKLQGLLKSA 770
Query 109 SLRVPGRHALNSATIAAIGVLGAL-FCVSSGHLTRMLCLYVNAGLSMWLGFHLVAAI 164
L +PGRH LN+ +AA V G + F V T + CL + LS +G L AAI
Sbjct 771 PLLLPGRHLLNAGLLAA-SVGGIIPFMVDPSFTTGITCLGSVSALSAVMGVTLTAAI 826
> xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydrogenase
(EC:1.6.1.2); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1086
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 55/168 (32%), Positives = 92/168 (54%), Gaps = 19/168 (11%)
Query 3 LMAEGVGAAAPQGAVQQTTAD--EVAD--ELLAARKVLIVPGYGMA-VARCQGGLAAVMV 57
L+A+ GA A G + T A +++D +L+AA L+ G+A V C +A M+
Sbjct 672 LLAQMSGAMALGGTIGLTIAKRIQISDLPQLVAAFHSLV----GLAAVLTC---VAEYMI 724
Query 58 GFANFHSPAGVERASSLLRLLEVYAGVFVAGITFTGSVVAAAKLHGSMESRSLRVPGRHA 117
+ +F + + A++L +++ Y G ++ G+TF+GS+VA KL G + S L +PGRH
Sbjct 725 EYPHFAT----DPAANLTKIV-AYLGTYIGGVTFSGSLVAYGKLQGVLNSAPLLLPGRHM 779
Query 118 LNSATIAAIGVLGAL-FCVSSGHLTRMLCLYVNAGLSMWLGFHLVAAI 164
LN+ + A V G + + + + T + CL + LS +G L AAI
Sbjct 780 LNAGLLTA-SVGGIIPYMLDPSYTTGITCLGSVSALSAVMGVTLTAAI 826
> dre:100331743 Nicotinamide Nucleotide Transhydrogenase family
member (nnt-1)-like
Length=518
Score = 55.8 bits (133), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 40/107 (37%), Positives = 63/107 (58%), Gaps = 6/107 (5%)
Query 51 GLAAVMVGFANF---HSPAGVERASSLLRLLEVYAGVFVAGITFTGSVVAAAKLHGSMES 107
GLAAV+ A + + V A+ +L+++ Y G ++ G+TF+GS+VA KL G ++S
Sbjct 411 GLAAVLTCVAEYMIEYPHLDVHPAAGVLKIV-AYLGTYIGGVTFSGSLVAYGKLQGLLDS 469
Query 108 RSLRVPGRHALNSA-TIAAIGVLGALFCVSSGHLTRMLCLYVNAGLS 153
L +PGRH LN+ +A++G + F +S T M CL +GLS
Sbjct 470 APLLLPGRHMLNAGLMLASVGGM-VPFMLSDSFHTGMGCLLGVSGLS 515
> cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase
family member (nnt-1); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1041
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 43/118 (36%), Positives = 58/118 (49%), Gaps = 5/118 (4%)
Query 51 GLAAVMVGFANF--HSPAGVERASSLLRLLEVYA-GVFVAGITFTGSVVAAAKLHGSMES 107
GLAA + ANF P +E S+ G ++ G+TFTGS++A KL G + S
Sbjct 666 GLAATLTCLANFIQEHPHFLEDPSNAAAAKLALFLGTYIGGVTFTGSLMAYGKLQGILAS 725
Query 108 RSLRVPGRHALNSATIAA-IGVLGALFCVSSGHLTRMLCLYVNAGLSMWLGFHLVAAI 164
+P RH LN A +A +G LG + S+ T M L GLS +G L AI
Sbjct 726 APTYLPARHVLNGALLAGNVGALGT-YMYSTDFGTGMSMLGGTVGLSSLMGVTLTMAI 782
Score = 39.3 bits (90), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 20/46 (43%), Positives = 29/46 (63%), Gaps = 1/46 (2%)
Query 5 AEGVG-AAAPQGAVQQTTADEVADELLAARKVLIVPGYGMAVARCQ 49
++G G A A +G ++ E AD LL AR V+I+PGYG+ A+ Q
Sbjct 852 SKGTGEAKAIEGTAKEIAPVETADMLLNARSVIIIPGYGLCAAQAQ 897
> tgo:TGME49_101210 NAD(P) transhydrogenase, alpha subunit, putative
(EC:1.6.1.2)
Length=1165
Score = 39.3 bits (90), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 20/41 (48%), Positives = 29/41 (70%), Gaps = 0/41 (0%)
Query 9 GAAAPQGAVQQTTADEVADELLAARKVLIVPGYGMAVARCQ 49
A A +G + T+++VA LLA++ V+IVPGYGMAV+ Q
Sbjct 268 NAQAFEGEAKIATSEQVAAYLLASQSVIIVPGYGMAVSHAQ 308
Score = 38.9 bits (89), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 40/79 (50%), Gaps = 13/79 (16%)
Query 50 GGLAAVMVGFANFHSPAGVERASSLLRL-------LEVYAGVF------VAGITFTGSVV 96
GGLAA + FA + SP V + ++ + +VY VF + ITFTGS+V
Sbjct 65 GGLAATLESFALYFSPYEVAKQEAMQSVGETRYLQFQVYQTVFYLIGASIGMITFTGSLV 124
Query 97 AAAKLHGSMESRSLRVPGR 115
A KL G + S+ +P R
Sbjct 125 ACGKLSGWIASKPRVMPLR 143
> bbo:BBOV_II006920 18.m06571; citrate synthase (EC:2.3.3.1);
K01647 citrate synthase [EC:2.3.3.1]
Length=476
Score = 32.3 bits (72), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 22/82 (26%), Positives = 37/82 (45%), Gaps = 7/82 (8%)
Query 14 QGAVQQTTADEVADELLAARKVLIVPGYGMAVARCQGGLAAVMVGFANFHSPAGVERASS 73
+G + T + A + L +K ++PGYG AV R + V FA+ H P
Sbjct 332 KGNITLETVTKFAQDTL--KKGQVIPGYGHAVLRVEDPRHTAFVNFAHKHFP-----DDP 384
Query 74 LLRLLEVYAGVFVAGITFTGSV 95
L+++L++ + TG V
Sbjct 385 LVKVLDICLKAIPPVLEATGKV 406
> dre:560948 fi40d06, wu:fi40d06; zgc:174160
Length=896
Score = 30.0 bits (66), Expect = 4.0, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 33/64 (51%), Gaps = 3/64 (4%)
Query 43 MAVARCQGGLAAVMVGFANFHSPAGVERASSLLRLLEVYAGVFVAGITFTGSVVAAAKLH 102
M V+ C L ++++ + + AS L + +E +G+ AG+ F+G V+ + L
Sbjct 1 MDVSTC---LKSLLLAIRQYRNNRSALNASQLQKHIEDISGLKCAGVLFSGQVLPSECLS 57
Query 103 GSME 106
G ME
Sbjct 58 GLME 61
> ath:AT5G42430 F-box family protein
Length=395
Score = 29.6 bits (65), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 12/25 (48%), Positives = 18/25 (72%), Gaps = 0/25 (0%)
Query 76 RLLEVYAGVFVAGITFTGSVVAAAK 100
R+++VY +F+ GIT TG +V A K
Sbjct 299 RVVKVYYDLFIVGITATGEIVLAKK 323
> hsa:92211 CDHR1, CORD15, DKFZp434A132, KIAA1775, PCDH21, PRCAD;
cadherin-related family member 1
Length=745
Score = 29.6 bits (65), Expect = 5.2, Method: Composition-based stats.
Identities = 23/83 (27%), Positives = 36/83 (43%), Gaps = 9/83 (10%)
Query 4 MAEGVGAAAPQGAVQQTTADEVADELLAARKVLIVPG---------YGMAVARCQGGLAA 54
+++G+ A + + T A++ A + V +VP AV R G +
Sbjct 110 ISDGLNLVAEKVVILVTDANDEAPRFIQEPYVALVPEDIPAGSIIFKVHAVDRDTGSGGS 169
Query 55 VMVGFANFHSPAGVERASSLLRL 77
V N HSP V+R S +LRL
Sbjct 170 VTYFLQNLHSPFAVDRHSGVLRL 192
> hsa:259287 TAS2R41, MGC119928, MGC119929, T2R41, T2R59; taste
receptor, type 2, member 41; K08474 taste receptor type 2
Length=307
Score = 29.3 bits (64), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 24/49 (48%), Gaps = 0/49 (0%)
Query 116 HALNSATIAAIGVLGALFCVSSGHLTRMLCLYVNAGLSMWLGFHLVAAI 164
H LNSAT L LFCV ++T L++ W+ + L+ ++
Sbjct 90 HFLNSATFWFCSWLSVLFCVKIANITHSTFLWLKWRFPGWVPWLLLGSV 138
Lambda K H
0.324 0.135 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4092719652
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40