bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
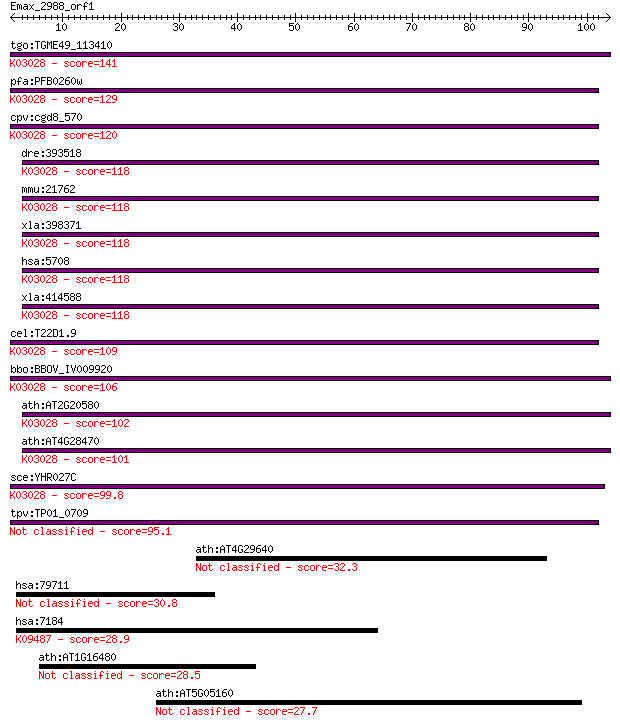
Query= Emax_2988_orf1
Length=103
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_113410 26S proteasome non-ATPase regulatory subunit... 141 4e-34
pfa:PFB0260w proteasome 26S regulatory subunit, putative; K030... 129 2e-30
cpv:cgd8_570 proteasome regulatory subunit S2 (RPN1) ; K03028 ... 120 8e-28
dre:393518 psmd2, MGC65921, zgc:65921; proteasome (prosome, ma... 118 4e-27
mmu:21762 Psmd2, 9430095H01Rik, AA407121, TEG-190, Tex190; pro... 118 4e-27
xla:398371 psmd2, MGC83116; proteasome (prosome, macropain) 26... 118 5e-27
hsa:5708 PSMD2, MGC14274, P97, Rpn1, S2, TRAP2; proteasome (pr... 118 5e-27
xla:414588 psmd2, MGC83233; proteasome (prosome, macropain) 26... 118 5e-27
cel:T22D1.9 rpn-1; proteasome Regulatory Particle, Non-ATPase-... 109 2e-24
bbo:BBOV_IV009920 23.m05991; proteasome 26S regulatory subunit... 106 2e-23
ath:AT2G20580 RPN1A; RPN1A (26S PROTEASOME REGULATORY SUBUNIT ... 102 3e-22
ath:AT4G28470 RPN1B; RPN1B (26S PROTEASOME REGULATORY SUBUNIT ... 101 7e-22
sce:YHR027C RPN1, HRD2, NAS1; Non-ATPase base subunit of the 1... 99.8 2e-21
tpv:TP01_0709 26S proteasome regulatory subunit 2 95.1
ath:AT4G29640 cytidine deaminase, putative / cytidine aminohyd... 32.3 0.35
hsa:79711 IPO4, FLJ23338, Imp4, MGC131665; importin 4 30.8
hsa:7184 HSP90B1, ECGP, GP96, GRP94, TRA1; heat shock protein ... 28.9 3.8
ath:AT1G16480 hypothetical protein 28.5 6.0
ath:AT5G05160 leucine-rich repeat transmembrane protein kinase... 27.7 9.7
> tgo:TGME49_113410 26S proteasome non-ATPase regulatory subunit
2, putative ; K03028 26S proteasome regulatory subunit N1
Length=1091
Score = 141 bits (356), Expect = 4e-34, Method: Composition-based stats.
Identities = 71/103 (68%), Positives = 86/103 (83%), Gaps = 0/103 (0%)
Query 1 PQLIDLLSKLSHDADPDTALNAIFAMGILGAGTNHSRIASLLRQLASYYGKDPSALFVVR 60
P +ID LSKLSHDAD D ALNAIFAMG++GAGTN++RIA LLR LASYYG+D +ALFV+R
Sbjct 837 PGVIDTLSKLSHDADADVALNAIFAMGVVGAGTNNARIAGLLRSLASYYGRDGNALFVIR 896
Query 61 LSQGLLYLGKGLLHIGALHSDRQLICRVALGALAIVSYSALLM 103
L+QGLLY+GKGLL I LHSDR LI VALG L+++ ++ L M
Sbjct 897 LAQGLLYMGKGLLTINPLHSDRFLINNVALGCLSVIMHTCLHM 939
> pfa:PFB0260w proteasome 26S regulatory subunit, putative; K03028
26S proteasome regulatory subunit N1
Length=966
Score = 129 bits (325), Expect = 2e-30, Method: Composition-based stats.
Identities = 61/101 (60%), Positives = 83/101 (82%), Gaps = 0/101 (0%)
Query 1 PQLIDLLSKLSHDADPDTALNAIFAMGILGAGTNHSRIASLLRQLASYYGKDPSALFVVR 60
P ++D+LSKL+HD DPD AL+AI ++G +GAGTN+SRIA LLRQL+++Y KD +A+FVVR
Sbjct 758 PNIVDILSKLTHDQDPDVALHAIISLGFVGAGTNNSRIAILLRQLSAFYCKDTNAIFVVR 817
Query 61 LSQGLLYLGKGLLHIGALHSDRQLICRVALGALAIVSYSAL 101
L+QGLLY+GKGLL I LHS+R +I V+LG+L I ++ L
Sbjct 818 LAQGLLYMGKGLLTINPLHSNRSIINYVSLGSLLITIHACL 858
> cpv:cgd8_570 proteasome regulatory subunit S2 (RPN1) ; K03028
26S proteasome regulatory subunit N1
Length=1045
Score = 120 bits (302), Expect = 8e-28, Method: Composition-based stats.
Identities = 56/101 (55%), Positives = 79/101 (78%), Gaps = 0/101 (0%)
Query 1 PQLIDLLSKLSHDADPDTALNAIFAMGILGAGTNHSRIASLLRQLASYYGKDPSALFVVR 60
P LID +SKL+HD+DPD ALNAIFA+G++ AGTN+SR A+LLRQLASYYGKD AL+ VR
Sbjct 833 PVLIDTISKLTHDSDPDVALNAIFALGLISAGTNNSRTANLLRQLASYYGKDKHALYAVR 892
Query 61 LSQGLLYLGKGLLHIGALHSDRQLICRVALGALAIVSYSAL 101
++QG LY+GKGL+ + H+++ L+ ++L ++ + AL
Sbjct 893 IAQGFLYMGKGLVTVNPCHANKLLLNPISLVSIITTLHLAL 933
> dre:393518 psmd2, MGC65921, zgc:65921; proteasome (prosome,
macropain) 26S subunit, non-ATPase, 2; K03028 26S proteasome
regulatory subunit N1
Length=897
Score = 118 bits (296), Expect = 4e-27, Method: Composition-based stats.
Identities = 56/99 (56%), Positives = 78/99 (78%), Gaps = 0/99 (0%)
Query 3 LIDLLSKLSHDADPDTALNAIFAMGILGAGTNHSRIASLLRQLASYYGKDPSALFVVRLS 62
++D LSK SHDADP+ + N+IFAMG++G+GTN++R+A++LRQLA Y+ KDP+ LF+VRL+
Sbjct 695 ILDTLSKFSHDADPEVSHNSIFAMGLVGSGTNNARLAAMLRQLAQYHAKDPNNLFMVRLA 754
Query 63 QGLLYLGKGLLHIGALHSDRQLICRVALGALAIVSYSAL 101
QGL +LGKG L + HSDRQL+ +VA+ L V S L
Sbjct 755 QGLTHLGKGTLTLCPYHSDRQLMSQVAVAGLLTVLVSFL 793
> mmu:21762 Psmd2, 9430095H01Rik, AA407121, TEG-190, Tex190; proteasome
(prosome, macropain) 26S subunit, non-ATPase, 2; K03028
26S proteasome regulatory subunit N1
Length=908
Score = 118 bits (296), Expect = 4e-27, Method: Composition-based stats.
Identities = 56/99 (56%), Positives = 78/99 (78%), Gaps = 0/99 (0%)
Query 3 LIDLLSKLSHDADPDTALNAIFAMGILGAGTNHSRIASLLRQLASYYGKDPSALFVVRLS 62
++D LSK SHDADP+ + N+IFAMG++G+GTN++R+A++LRQLA Y+ KDP+ LF+VRL+
Sbjct 706 ILDTLSKFSHDADPEVSYNSIFAMGMVGSGTNNARLAAMLRQLAQYHAKDPNNLFMVRLA 765
Query 63 QGLLYLGKGLLHIGALHSDRQLICRVALGALAIVSYSAL 101
QGL +LGKG L + HSDRQL+ +VA+ L V S L
Sbjct 766 QGLTHLGKGTLTLCPYHSDRQLMSQVAVAGLLTVLVSFL 804
> xla:398371 psmd2, MGC83116; proteasome (prosome, macropain)
26S subunit, non-ATPase, 2; K03028 26S proteasome regulatory
subunit N1
Length=897
Score = 118 bits (296), Expect = 5e-27, Method: Composition-based stats.
Identities = 56/99 (56%), Positives = 78/99 (78%), Gaps = 0/99 (0%)
Query 3 LIDLLSKLSHDADPDTALNAIFAMGILGAGTNHSRIASLLRQLASYYGKDPSALFVVRLS 62
++D LSK SHDADP+ + N+IFAMG++G+GTN++R+A++LRQLA Y+ KDP+ LF+VRL+
Sbjct 695 ILDTLSKFSHDADPEVSYNSIFAMGMVGSGTNNARLAAMLRQLAQYHAKDPNNLFMVRLA 754
Query 63 QGLLYLGKGLLHIGALHSDRQLICRVALGALAIVSYSAL 101
QGL +LGKG L + HSDRQL+ +VA+ L V S L
Sbjct 755 QGLTHLGKGTLTLCPYHSDRQLMSQVAVAGLLTVLVSFL 793
> hsa:5708 PSMD2, MGC14274, P97, Rpn1, S2, TRAP2; proteasome (prosome,
macropain) 26S subunit, non-ATPase, 2; K03028 26S proteasome
regulatory subunit N1
Length=908
Score = 118 bits (296), Expect = 5e-27, Method: Composition-based stats.
Identities = 56/99 (56%), Positives = 78/99 (78%), Gaps = 0/99 (0%)
Query 3 LIDLLSKLSHDADPDTALNAIFAMGILGAGTNHSRIASLLRQLASYYGKDPSALFVVRLS 62
++D LSK SHDADP+ + N+IFAMG++G+GTN++R+A++LRQLA Y+ KDP+ LF+VRL+
Sbjct 706 ILDTLSKFSHDADPEVSYNSIFAMGMVGSGTNNARLAAMLRQLAQYHAKDPNNLFMVRLA 765
Query 63 QGLLYLGKGLLHIGALHSDRQLICRVALGALAIVSYSAL 101
QGL +LGKG L + HSDRQL+ +VA+ L V S L
Sbjct 766 QGLTHLGKGTLTLCPYHSDRQLMSQVAVAGLLTVLVSFL 804
> xla:414588 psmd2, MGC83233; proteasome (prosome, macropain)
26S subunit, non-ATPase, 2; K03028 26S proteasome regulatory
subunit N1
Length=897
Score = 118 bits (295), Expect = 5e-27, Method: Composition-based stats.
Identities = 56/99 (56%), Positives = 78/99 (78%), Gaps = 0/99 (0%)
Query 3 LIDLLSKLSHDADPDTALNAIFAMGILGAGTNHSRIASLLRQLASYYGKDPSALFVVRLS 62
++D LSK SHDADP+ + N+IFAMG++G+GTN++R+A++LRQLA Y+ KDP+ LF+VRL+
Sbjct 695 ILDTLSKFSHDADPEVSYNSIFAMGMVGSGTNNARLAAMLRQLAQYHAKDPNNLFMVRLA 754
Query 63 QGLLYLGKGLLHIGALHSDRQLICRVALGALAIVSYSAL 101
QGL +LGKG L + HSDRQL+ +VA+ L V S L
Sbjct 755 QGLTHLGKGTLTLCPYHSDRQLMSQVAVAGLLTVLVSFL 793
> cel:T22D1.9 rpn-1; proteasome Regulatory Particle, Non-ATPase-like
family member (rpn-1); K03028 26S proteasome regulatory
subunit N1
Length=981
Score = 109 bits (273), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 55/103 (53%), Positives = 77/103 (74%), Gaps = 2/103 (1%)
Query 1 PQL--IDLLSKLSHDADPDTALNAIFAMGILGAGTNHSRIASLLRQLASYYGKDPSALFV 58
PQL ++ LSK SHD+D DTA NAIFAMG++GAGTN++R+ ++LR LASY+ KD +L +
Sbjct 761 PQLNILETLSKFSHDSDADTAHNAIFAMGLVGAGTNNARLVAMLRNLASYHYKDQVSLML 820
Query 59 VRLSQGLLYLGKGLLHIGALHSDRQLICRVALGALAIVSYSAL 101
VR++QGL +LGKG + + HSDRQL+ AL +L + +S L
Sbjct 821 VRIAQGLTHLGKGTMTLNPWHSDRQLLSPSALASLLSICFSFL 863
> bbo:BBOV_IV009920 23.m05991; proteasome 26S regulatory subunit;
K03028 26S proteasome regulatory subunit N1
Length=1008
Score = 106 bits (265), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 55/103 (53%), Positives = 73/103 (70%), Gaps = 0/103 (0%)
Query 1 PQLIDLLSKLSHDADPDTALNAIFAMGILGAGTNHSRIASLLRQLASYYGKDPSALFVVR 60
PQ+ID+LSKL+HDAD +NAIF MGI+GAGTN+SRIA LL LA + +D F++R
Sbjct 802 PQVIDILSKLTHDADYQVTINAIFGMGIVGAGTNNSRIAVLLLNLAKTHARDSKGTFIIR 861
Query 61 LSQGLLYLGKGLLHIGALHSDRQLICRVALGALAIVSYSALLM 103
++ GLL++GKG + I LHS+ LI + AL L IV +AL M
Sbjct 862 IAAGLLHMGKGTMTISPLHSEGFLIRKPALAGLFIVIVAALDM 904
> ath:AT2G20580 RPN1A; RPN1A (26S PROTEASOME REGULATORY SUBUNIT
S2 1A); binding / enzyme regulator; K03028 26S proteasome
regulatory subunit N1
Length=891
Score = 102 bits (254), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 49/101 (48%), Positives = 75/101 (74%), Gaps = 0/101 (0%)
Query 3 LIDLLSKLSHDADPDTALNAIFAMGILGAGTNHSRIASLLRQLASYYGKDPSALFVVRLS 62
++D LS+LSHD D + A++AI ++G++GAGTN++RIA +LR L+SYY KD S LF VR++
Sbjct 688 VMDTLSRLSHDTDSEVAMSAIISLGLIGAGTNNARIAGMLRNLSSYYYKDMSLLFCVRIA 747
Query 63 QGLLYLGKGLLHIGALHSDRQLICRVALGALAIVSYSALLM 103
QGL+++GKGLL + HS+R L+ AL + + ++ L M
Sbjct 748 QGLVHMGKGLLTLSPFHSERFLLSPTALAGIVTLLHACLDM 788
> ath:AT4G28470 RPN1B; RPN1B (26S PROTEASOME REGULATORY SUBUNIT
S2 1B); binding / enzyme regulator; K03028 26S proteasome
regulatory subunit N1
Length=891
Score = 101 bits (251), Expect = 7e-22, Method: Compositional matrix adjust.
Identities = 48/101 (47%), Positives = 73/101 (72%), Gaps = 0/101 (0%)
Query 3 LIDLLSKLSHDADPDTALNAIFAMGILGAGTNHSRIASLLRQLASYYGKDPSALFVVRLS 62
++D LS+LSHD D + A+ AI ++G++GAGTN++RIA +LR L+SYY KD S LF VR++
Sbjct 688 VMDTLSRLSHDTDSEVAMAAIISLGLIGAGTNNARIAGMLRNLSSYYYKDASLLFCVRIA 747
Query 63 QGLLYLGKGLLHIGALHSDRQLICRVALGALAIVSYSALLM 103
QG +++GKGLL + HS+R L+ AL + + ++ L M
Sbjct 748 QGFVHMGKGLLTLNPFHSERLLLSPTALAGIVTLLHACLDM 788
> sce:YHR027C RPN1, HRD2, NAS1; Non-ATPase base subunit of the
19S regulatory particle of the 26S proteasome; may participate
in the recognition of several ligands of the proteasome;
contains a leucine-rich repeat (LRR) domain, a site for protein-protein
interactions; K03028 26S proteasome regulatory
subunit N1
Length=993
Score = 99.8 bits (247), Expect = 2e-21, Method: Composition-based stats.
Identities = 52/109 (47%), Positives = 77/109 (70%), Gaps = 8/109 (7%)
Query 1 PQL--IDLLSKLSHDADPDTALNAIFAMGILGAGTNHSRIASLLRQLASYYGKDPSALFV 58
PQ+ D L++ SHDAD + ++N+IFAMG+ GAGTN++R+A LLRQLASYY ++ ALF+
Sbjct 788 PQMKVFDTLTRFSHDADLEVSMNSIFAMGLCGAGTNNARLAQLLRQLASYYSREQDALFI 847
Query 59 VRLSQGLLYLGKGLLHIGALHSDRQLICRVALG-----ALAIVSYSALL 102
RL+QGLL+LGKG + + +D ++ +V L A+ +VS S +L
Sbjct 848 TRLAQGLLHLGKGTMTMDVF-NDAHVLNKVTLASILTTAVGLVSPSFML 895
> tpv:TP01_0709 26S proteasome regulatory subunit 2
Length=1123
Score = 95.1 bits (235), Expect = 5e-20, Method: Composition-based stats.
Identities = 46/101 (45%), Positives = 71/101 (70%), Gaps = 0/101 (0%)
Query 1 PQLIDLLSKLSHDADPDTALNAIFAMGILGAGTNHSRIASLLRQLASYYGKDPSALFVVR 60
P ++ LSKLSHD+D D +L AI +G++GAGTN+SRI+ LL+ +A + +D + ++V+R
Sbjct 920 PAIVSALSKLSHDSDKDVSLCAILGLGLVGAGTNNSRISQLLQNIAKHSYRDSTLMYVLR 979
Query 61 LSQGLLYLGKGLLHIGALHSDRQLICRVALGALAIVSYSAL 101
LS GLL++GKG L + LHSD ++ + +L L I + AL
Sbjct 980 LSVGLLFMGKGTLTLTPLHSDSFILNKTSLAGLLITLFCAL 1020
> ath:AT4G29640 cytidine deaminase, putative / cytidine aminohydrolase,
putative
Length=346
Score = 32.3 bits (72), Expect = 0.35, Method: Composition-based stats.
Identities = 24/66 (36%), Positives = 37/66 (56%), Gaps = 7/66 (10%)
Query 33 TNHSRIASLLRQLASYYGKDPSALFVV-RLSQGLLYLG-----KGLLHIGALHSDRQLIC 86
T+H ++ L+R+ A K PS + V R S G YLG KGLL ++H+++ LI
Sbjct 20 TDHKKLPKLIRK-ARNLVKAPSKVGAVGRASSGRFYLGVNVEFKGLLPHFSIHAEQFLIA 78
Query 87 RVALGA 92
+AL +
Sbjct 79 NLALNS 84
> hsa:79711 IPO4, FLJ23338, Imp4, MGC131665; importin 4
Length=1081
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 13/34 (38%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 2 QLIDLLSKLSHDADPDTALNAIFAMGILGAGTNH 35
+L+ +L + +ADP+ NAIF MG+L H
Sbjct 899 RLLPVLLSTAQEADPEVRSNAIFGMGVLAEHGGH 932
> hsa:7184 HSP90B1, ECGP, GP96, GRP94, TRA1; heat shock protein
90kDa beta (Grp94), member 1; K09487 heat shock protein 90kDa
beta
Length=803
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 17/67 (25%), Positives = 37/67 (55%), Gaps = 5/67 (7%)
Query 2 QLIDLLSKLSHDADPDT---ALNAIFAMGILGAGTNHSRIASLLRQLASYYGKDPSAL-- 56
+ +D++ K++ D DT +G++ +N +R+A LLR +S++ D ++L
Sbjct 467 KTLDMIKKIADDKYNDTFWKEFGTNIKLGVIEDHSNRTRLAKLLRFQSSHHPTDITSLDQ 526
Query 57 FVVRLSQ 63
+V R+ +
Sbjct 527 YVERMKE 533
> ath:AT1G16480 hypothetical protein
Length=937
Score = 28.5 bits (62), Expect = 6.0, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 6 LLSKLSHDADPDTALNAIFAMGILGAGTNHSRIASLL 42
L+ + D DPD AL A M + G +N+ + S+L
Sbjct 434 LIGGYAEDEDPDKALAAFQTMRVEGVSSNYITVVSVL 470
> ath:AT5G05160 leucine-rich repeat transmembrane protein kinase,
putative
Length=640
Score = 27.7 bits (60), Expect = 9.7, Method: Composition-based stats.
Identities = 23/75 (30%), Positives = 34/75 (45%), Gaps = 8/75 (10%)
Query 26 MGILGAGTNHSRIASLLRQLASYYGKDPSALFVVRLSQGLLYLGKGLLH--IGALHSDRQ 83
M I+G HS LL A YY KD L +++G L+ G++H G D +
Sbjct 391 MEIVGKINQHSNFVPLL---AYYYSKDEKLLVYKYMTKGSLF---GIMHGNRGDRGVDWE 444
Query 84 LICRVALGALAIVSY 98
++A G +SY
Sbjct 445 TRMKIATGTSKAISY 459
Lambda K H
0.325 0.140 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2027061836
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40