bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2968_orf1
Length=133
Score E
Sequences producing significant alignments: (Bits) Value
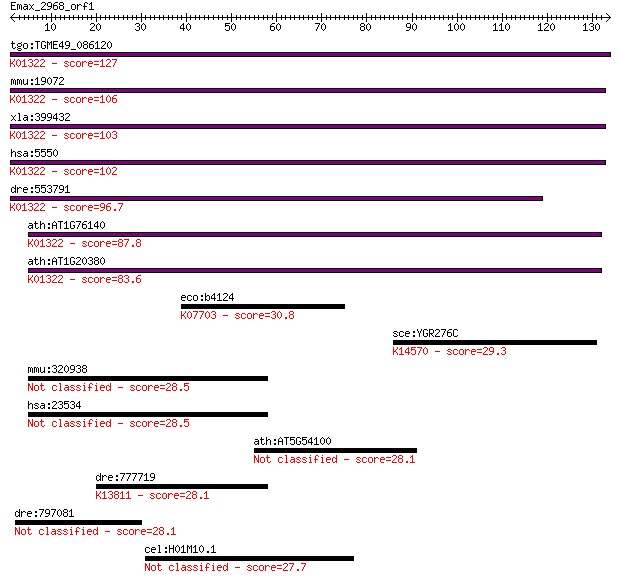
tgo:TGME49_086120 prolyl endopeptidase, putative (EC:3.4.21.26... 127 1e-29
mmu:19072 Prep, AI047692, AI450383, D10Wsu136e, PEP, Pop; prol... 106 2e-23
xla:399432 prep, xprep; prolyl endopeptidase (EC:3.4.21.26); K... 103 2e-22
hsa:5550 PREP, MGC16060, PE, PEP; prolyl endopeptidase (EC:3.4... 102 2e-22
dre:553791 prep, MGC110670, im:7140031, zgc:110670; prolyl end... 96.7 2e-20
ath:AT1G76140 serine-type endopeptidase/ serine-type peptidase... 87.8 8e-18
ath:AT1G20380 prolyl oligopeptidase, putative / prolyl endopep... 83.6 1e-16
eco:b4124 dcuR, ECK4117, JW4085, yjdG; DNA-binding response re... 30.8 1.2
sce:YGR276C RNH70, REX1, RNA82; 3'-5' exoribonuclease; require... 29.3 3.7
mmu:320938 Tnpo3, 5730544L10Rik, C430013M08Rik, C81142, D6Ertd... 28.5 6.2
hsa:23534 TNPO3, IPO12, MTR10A, TRN-SR, TRN-SR2, TRNSR; transp... 28.5 6.2
ath:AT5G54100 band 7 family protein 28.1 7.7
dre:777719 papss2a, zgc:153748; 3'-phosphoadenosine 5'-phospho... 28.1 8.2
dre:797081 si:ch211-152b13.3, wu:fc56e12; si:ch211-152b13.4 28.1 8.6
cel:H01M10.1 hypothetical protein 27.7 8.9
> tgo:TGME49_086120 prolyl endopeptidase, putative (EC:3.4.21.26);
K01322 prolyl oligopeptidase [EC:3.4.21.26]
Length=825
Score = 127 bits (318), Expect = 1e-29, Method: Composition-based stats.
Identities = 63/139 (45%), Positives = 92/139 (66%), Gaps = 7/139 (5%)
Query 1 FPSEPTWMVSVDVTDDEKFIVASISRSCEPTNKLWIAALPDGTVPSTA-----FSKLQWV 55
FP +P W+ + V+ D +F++A I+ SC PTN++WIA L D S A FSKL+W
Sbjct 331 FPDQPKWLSEITVSWDHRFLIAGIAESCSPTNQVWIADL-DTIRTSNAQGMFDFSKLKWN 389
Query 56 KVADDFEAGFDYVANDGTIFLLKTNRDAPKNKIVKVDVAKP-DAPMVDVVPEAGAVLEDV 114
K+ ++F+AG++Y+ NDGT F+ TN+DAP K+VK+D+AKP +A ++ E +L
Sbjct 390 KIIENFDAGYEYLTNDGTEFVFLTNKDAPLYKLVKIDLAKPEEASWKTLIEERTELLSSA 449
Query 115 MVVAGDKMVLHYNEDVKSK 133
VV GDK+VLHY DV S+
Sbjct 450 DVVDGDKLVLHYFRDVTSQ 468
> mmu:19072 Prep, AI047692, AI450383, D10Wsu136e, PEP, Pop; prolyl
endopeptidase (EC:3.4.21.26); K01322 prolyl oligopeptidase
[EC:3.4.21.26]
Length=710
Score = 106 bits (264), Expect = 2e-23, Method: Composition-based stats.
Identities = 54/134 (40%), Positives = 79/134 (58%), Gaps = 4/134 (2%)
Query 1 FPSEPTWMVSVDVTDDEKFIVASISRSCEPTNKLWIAALPDGTVPSTAFSKLQWVKVADD 60
FP EP WM +++DD ++++ SI C+P N+LW L P+ L+WVK+ D+
Sbjct 228 FPDEPKWMGGAELSDDGRYVLLSIWEGCDPVNRLWYCDLQQE--PNGITGILKWVKLIDN 285
Query 61 FEAGFDYVANDGTIFLLKTNRDAPKNKIVKVDVAKPDAPMVDV-VPE-AGAVLEDVMVVA 118
FE +DYV N+GT+F KTNR++P +++ +D PD V VPE VLE V V
Sbjct 286 FEGEYDYVTNEGTVFTFKTNRNSPNYRLINIDFTDPDESKWKVLVPEHEKDVLEWVACVR 345
Query 119 GDKMVLHYNEDVKS 132
+ +VL Y DVK+
Sbjct 346 SNFLVLCYLHDVKN 359
> xla:399432 prep, xprep; prolyl endopeptidase (EC:3.4.21.26);
K01322 prolyl oligopeptidase [EC:3.4.21.26]
Length=712
Score = 103 bits (256), Expect = 2e-22, Method: Composition-based stats.
Identities = 55/134 (41%), Positives = 78/134 (58%), Gaps = 5/134 (3%)
Query 1 FPSEPTWMVSVDVTDDEKFIVASISRSCEPTNKLWIAALPDGTVPSTAFSKLQWVKVADD 60
FP EP WM +VTDD ++++ SI C+P N+LW L T + L WVK+ D+
Sbjct 231 FPEEPKWMGGAEVTDDGQYVLLSIREGCDPVNRLWYCKLNKNTGIT---GTLPWVKLIDN 287
Query 61 FEAGFDYVANDGTIFLLKTNRDAPKNKIVKVDVAKP-DAPMVDVVPE-AGAVLEDVMVVA 118
FEA ++Y+ N+GTIF KTNR+AP ++ +D P ++ +VPE VLE V V
Sbjct 288 FEAEYEYITNEGTIFTFKTNRNAPNYNLINIDFNNPEESNWKALVPEHQKDVLEWVSCVH 347
Query 119 GDKMVLHYNEDVKS 132
+VL Y DVK+
Sbjct 348 KKFLVLCYLHDVKN 361
> hsa:5550 PREP, MGC16060, PE, PEP; prolyl endopeptidase (EC:3.4.21.26);
K01322 prolyl oligopeptidase [EC:3.4.21.26]
Length=710
Score = 102 bits (255), Expect = 2e-22, Method: Composition-based stats.
Identities = 52/134 (38%), Positives = 78/134 (58%), Gaps = 4/134 (2%)
Query 1 FPSEPTWMVSVDVTDDEKFIVASISRSCEPTNKLWIAALPDGTVPSTAFSKLQWVKVADD 60
FP EP WM +++DD ++++ SI C+P N+LW L + S L+WVK+ D+
Sbjct 228 FPDEPKWMGGAELSDDGRYVLLSIREGCDPVNRLWYCDLQQES--SGIAGILKWVKLIDN 285
Query 61 FEAGFDYVANDGTIFLLKTNRDAPKNKIVKVDVAKPDAPMVDV-VPE-AGAVLEDVMVVA 118
FE +DYV N+GT+F KTNR +P +++ +D P+ V VPE VLE + V
Sbjct 286 FEGEYDYVTNEGTVFTFKTNRQSPNYRVINIDFRDPEESKWKVLVPEHEKDVLEWIACVR 345
Query 119 GDKMVLHYNEDVKS 132
+ +VL Y DVK+
Sbjct 346 SNFLVLCYLHDVKN 359
> dre:553791 prep, MGC110670, im:7140031, zgc:110670; prolyl endopeptidase
(EC:3.4.21.26); K01322 prolyl oligopeptidase [EC:3.4.21.26]
Length=709
Score = 96.7 bits (239), Expect = 2e-20, Method: Composition-based stats.
Identities = 44/119 (36%), Positives = 73/119 (61%), Gaps = 6/119 (5%)
Query 1 FPSEPTWMVSVDVTDDEKFIVASISRSCEPTNKLWIAALPDGTVPSTAFSKLQWVKVADD 60
FP EP WM V+V+DD ++++ SI C+P N+LW L + P L WVK+ D+
Sbjct 227 FPDEPKWMSGVEVSDDGRYVLLSIREGCDPVNRLWYCDLNEE--PQGITGLLPWVKLIDN 284
Query 61 FEAGFDYVANDGTIFLLKTNRDAPKNKIVKVDVAKPDAPM-VDVVPEAGAVLEDVMVVA 118
F+A ++YV N+GT+F KTN +AP+ +++ +D +P +++P+ +DV+V A
Sbjct 285 FDAEYEYVTNEGTVFTFKTNLEAPQYRLINIDFTQPSVSQWKELIPQHD---KDVIVFA 340
> ath:AT1G76140 serine-type endopeptidase/ serine-type peptidase;
K01322 prolyl oligopeptidase [EC:3.4.21.26]
Length=792
Score = 87.8 bits (216), Expect = 8e-18, Method: Compositional matrix adjust.
Identities = 50/132 (37%), Positives = 77/132 (58%), Gaps = 6/132 (4%)
Query 5 PTWMVSVDVTDDEKFIVASISRSCEPTNKLWIAALPDGTVPSTAF----SKLQWVKVADD 60
P +M +VTDD K+++ SI SC+P NKL+ + + +F S L ++K+ D
Sbjct 306 PKYMFGAEVTDDGKYLIMSIGESCDPVNKLYYCDMTSLSGGLESFRGSSSFLPFIKLVDT 365
Query 61 FEAGFDYVANDGTIFLLKTNRDAPKNKIVKVDVAKPDAPMVDVVPEAGA-VLEDVMVVAG 119
F+A + ++ND T+F TN+DAPK K+V+VD+ +P++ DVV E VL V G
Sbjct 366 FDAQYSVISNDETLFTFLTNKDAPKYKLVRVDLKEPNS-WTDVVEEHEKDVLASACAVNG 424
Query 120 DKMVLHYNEDVK 131
+ +V Y DVK
Sbjct 425 NHLVACYMSDVK 436
> ath:AT1G20380 prolyl oligopeptidase, putative / prolyl endopeptidase,
putative / post-proline cleaving enzyme, putative;
K01322 prolyl oligopeptidase [EC:3.4.21.26]
Length=731
Score = 83.6 bits (205), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 51/132 (38%), Positives = 77/132 (58%), Gaps = 6/132 (4%)
Query 5 PTWMVSVDVTDDEKFIVASISRSCEPTNKLW---IAALPDGTVP-STAFSKLQWVKVADD 60
P M VTDD K+++ SI C+P NK++ ++ LP G + + L +VK+ D
Sbjct 242 PKHMFGSKVTDDGKYLIMSIEEGCDPVNKVYHCDLSLLPKGLEGFRGSNTLLPFVKLIDT 301
Query 61 FEAGFDYVANDGTIFLLKTNRDAPKNKIVKVDVAKPDAPMVDVVPEAGA-VLEDVMVVAG 119
F+A + +AND T+F TN+DAPK K+V+VD+ +P + DV+ E VL V G
Sbjct 302 FDAQYIAIANDETLFTFLTNKDAPKYKVVRVDLKEPSS-WTDVIAEHEKDVLSTASAVNG 360
Query 120 DKMVLHYNEDVK 131
D++V+ Y DVK
Sbjct 361 DQLVVSYMSDVK 372
> eco:b4124 dcuR, ECK4117, JW4085, yjdG; DNA-binding response
regulator in two-component regulatory system with DcuS; K07703
two-component system, CitB family, response regulator DcuR
Length=239
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 18/36 (50%), Gaps = 0/36 (0%)
Query 39 LPDGTVPSTAFSKLQWVKVADDFEAGFDYVANDGTI 74
LP G P T + QW+ D+E D +AN+ I
Sbjct 155 LPKGLTPQTLRTLCQWIDAHQDYEFSTDELANEVNI 190
> sce:YGR276C RNH70, REX1, RNA82; 3'-5' exoribonuclease; required
for maturation of 3' ends of 5S rRNA and tRNA-Arg3 from
dicistronic transcripts (EC:3.1.-.-); K14570 RNA exonuclease
1 [EC:3.1.-.-]
Length=553
Score = 29.3 bits (64), Expect = 3.7, Method: Composition-based stats.
Identities = 11/45 (24%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 86 NKIVKVDVAKPDAPMVDVVPEAGAVLEDVMVVAGDKMVLHYNEDV 130
N+++ ++ KPD P+VD + + E+ + V K + +D+
Sbjct 251 NEVIYEELVKPDVPIVDYLTRYSGITEEKLTVGAKKTLREVQKDL 295
> mmu:320938 Tnpo3, 5730544L10Rik, C430013M08Rik, C81142, D6Ertd313e,
KIAA4133, MGC90049, Trn-SR, mKIAA4133; transportin 3
Length=923
Score = 28.5 bits (62), Expect = 6.2, Method: Composition-based stats.
Identities = 15/55 (27%), Positives = 30/55 (54%), Gaps = 6/55 (10%)
Query 5 PTWMVSVDVTDDEKFIVASISRSCEPTNKLWIAALPDGTV--PSTAFSKLQWVKV 57
P W +VT+ FI+A+I++S +P N + + +G V P T + +++ +
Sbjct 432 PPW----EVTEAVLFIMAAIAKSVDPENNPTLVEVLEGVVHLPETVHTAVRYTSI 482
> hsa:23534 TNPO3, IPO12, MTR10A, TRN-SR, TRN-SR2, TRNSR; transportin
3
Length=859
Score = 28.5 bits (62), Expect = 6.2, Method: Composition-based stats.
Identities = 15/55 (27%), Positives = 30/55 (54%), Gaps = 6/55 (10%)
Query 5 PTWMVSVDVTDDEKFIVASISRSCEPTNKLWIAALPDGTV--PSTAFSKLQWVKV 57
P W +VT+ FI+A+I++S +P N + + +G V P T + +++ +
Sbjct 432 PPW----EVTEAVLFIMAAIAKSVDPENNPTLVEVLEGVVRLPETVHTAVRYTSI 482
> ath:AT5G54100 band 7 family protein
Length=401
Score = 28.1 bits (61), Expect = 7.7, Method: Composition-based stats.
Identities = 12/36 (33%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 55 VKVADDFEAGFDYVANDGTIFLLKTNRDAPKNKIVK 90
++VA+ + F +A +GT LL +N D P + I +
Sbjct 344 LRVAEQYIQAFGKIAKEGTTMLLPSNVDNPASMIAQ 379
> dre:777719 papss2a, zgc:153748; 3'-phosphoadenosine 5'-phosphosulfate
synthase 2a; K13811 3'-phosphoadenosine 5'-phosphosulfate
synthase [EC:2.7.7.4 2.7.1.25]
Length=612
Score = 28.1 bits (61), Expect = 8.2, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 23/40 (57%), Gaps = 2/40 (5%)
Query 20 IVASISRSCEPTNKLWIAALPDGTVPSTAFSK--LQWVKV 57
+V I+ P N+L +A T+PS + +K LQWV+V
Sbjct 217 VVEEINELFVPENRLKLAQAEASTLPSISITKLDLQWVQV 256
> dre:797081 si:ch211-152b13.3, wu:fc56e12; si:ch211-152b13.4
Length=2147
Score = 28.1 bits (61), Expect = 8.6, Method: Composition-based stats.
Identities = 15/28 (53%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 2 PSEPTWMVSVDVTDDEKFIVASISRSCE 29
PSE T +VS VTD KF +SI + CE
Sbjct 492 PSEITKVVSSKVTDGVKFSRSSIKQWCE 519
> cel:H01M10.1 hypothetical protein
Length=259
Score = 27.7 bits (60), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 24/46 (52%), Gaps = 1/46 (2%)
Query 31 TNKLWIAALPDGTVPSTAFSKLQWVKVADDFEAGFDYVANDGTIFL 76
NK I +LP VPST+ S Q + +E F VA DGTI +
Sbjct 61 NNKSRINSLPSSDVPSTSASCSQ-PRAPSVYEDPFVAVAADGTIHI 105
Lambda K H
0.316 0.132 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2165519004
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40