bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2881_orf2
Length=62
Score E
Sequences producing significant alignments: (Bits) Value
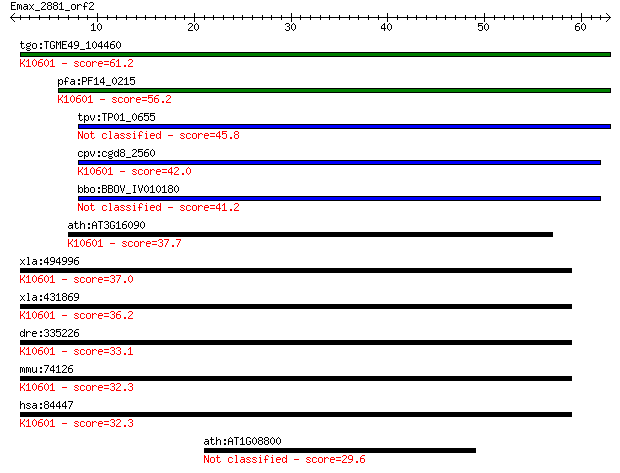
tgo:TGME49_104460 zinc finger (C3HC4 RING finger) protein, put... 61.2 8e-10
pfa:PF14_0215 conserved Plasmodium membrane protein, unknown f... 56.2 2e-08
tpv:TP01_0655 hypothetical protein 45.8 3e-05
cpv:cgd8_2560 HRD1 like membrane associated RING finger contai... 42.0 4e-04
bbo:BBOV_IV010180 23.m06036; zinc finger, C3HC4 type domain co... 41.2 0.001
ath:AT3G16090 zinc finger (C3HC4-type RING finger) family prot... 37.7 0.009
xla:494996 syvn1-b, hrd1; synovial apoptosis inhibitor 1, syno... 37.0 0.016
xla:431869 syvn1-a, MGC83718, hrd1; synovial apoptosis inhibit... 36.2 0.025
dre:335226 syvn1, wu:fk91f10, zgc:55735, zgc:77108; synovial a... 33.1 0.22
mmu:74126 Syvn1, 1200010C09Rik, AW211966, C85322, D530017H19Ri... 32.3 0.37
hsa:84447 SYVN1, HRD1, KIAA1810, MGC40372; synovial apoptosis ... 32.3 0.37
ath:AT1G08800 hypothetical protein 29.6 2.5
> tgo:TGME49_104460 zinc finger (C3HC4 RING finger) protein, putative
; K10601 E3 ubiquitin-protein ligase synoviolin [EC:6.3.2.19]
Length=710
Score = 61.2 bits (147), Expect = 8e-10, Method: Composition-based stats.
Identities = 28/61 (45%), Positives = 42/61 (68%), Gaps = 0/61 (0%)
Query 2 YLFNDEFYTAVVKLASNKLALAIEYNFCLLMFLLFVKFILHLLVGRLRRLEVEQVLDNGR 61
+ ++E YT V L++ K+ALA+ YN ++FL K + L+VG LR LE+EQV+D+GR
Sbjct 27 WYVHEELYTVVAVLSTGKIALALFYNCAFMLFLGLGKLAMRLMVGSLRDLEMEQVIDSGR 86
Query 62 G 62
G
Sbjct 87 G 87
> pfa:PF14_0215 conserved Plasmodium membrane protein, unknown
function; K10601 E3 ubiquitin-protein ligase synoviolin [EC:6.3.2.19]
Length=510
Score = 56.2 bits (134), Expect = 2e-08, Method: Composition-based stats.
Identities = 28/57 (49%), Positives = 40/57 (70%), Gaps = 0/57 (0%)
Query 6 DEFYTAVVKLASNKLALAIEYNFCLLMFLLFVKFILHLLVGRLRRLEVEQVLDNGRG 62
DEFY AVV L++ K I YNF L++F++ K +L++ +G LR LEVEQ++DN R
Sbjct 28 DEFYLAVVYLSTEKFPRTIIYNFFLMVFIVLCKLLLNVFIGELRYLEVEQLIDNARA 84
> tpv:TP01_0655 hypothetical protein
Length=618
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 35/55 (63%), Gaps = 0/55 (0%)
Query 8 FYTAVVKLASNKLALAIEYNFCLLMFLLFVKFILHLLVGRLRRLEVEQVLDNGRG 62
FY + + SNK LAI YN+CL++++ K + + +G L +LE E++++N R
Sbjct 31 FYDIITRFLSNKTCLAILYNYCLMLYIYCCKIPIFIFLGTLTQLESEELIENIRN 85
> cpv:cgd8_2560 HRD1 like membrane associated RING finger containing
protein signal peptide plus 6 transmembrane domains ;
K10601 E3 ubiquitin-protein ligase synoviolin [EC:6.3.2.19]
Length=637
Score = 42.0 bits (97), Expect = 4e-04, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 31/54 (57%), Gaps = 0/54 (0%)
Query 8 FYTAVVKLASNKLALAIEYNFCLLMFLLFVKFILHLLVGRLRRLEVEQVLDNGR 61
FY ++ + K + I N LL K IL++ VGRLR +E+E+++D GR
Sbjct 41 FYHIIMHITITKASTMIITNAGLLTLYFLGKVILNMFVGRLRDIELEEIIDQGR 94
> bbo:BBOV_IV010180 23.m06036; zinc finger, C3HC4 type domain
containing protein
Length=1151
Score = 41.2 bits (95), Expect = 0.001, Method: Composition-based stats.
Identities = 18/54 (33%), Positives = 32/54 (59%), Gaps = 0/54 (0%)
Query 8 FYTAVVKLASNKLALAIEYNFCLLMFLLFVKFILHLLVGRLRRLEVEQVLDNGR 61
FY +V SNK +A+ YN+ L+ F++ + + +GRL ++E EQ+ + R
Sbjct 34 FYEFIVFYMSNKTCVAVMYNYLLMWFIVMSILFVRIFLGRLSQMEREQLYEMTR 87
> ath:AT3G16090 zinc finger (C3HC4-type RING finger) family protein;
K10601 E3 ubiquitin-protein ligase synoviolin [EC:6.3.2.19]
Length=492
Score = 37.7 bits (86), Expect = 0.009, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 31/50 (62%), Gaps = 0/50 (0%)
Query 7 EFYTAVVKLASNKLALAIEYNFCLLMFLLFVKFILHLLVGRLRRLEVEQV 56
+FY A V L+++K++L + N CL++ L + + +G LR EVE++
Sbjct 28 QFYPATVYLSTSKISLVLLLNMCLVLMLSLWHLVKFVFLGSLREAEVERL 77
> xla:494996 syvn1-b, hrd1; synovial apoptosis inhibitor 1, synoviolin;
K10601 E3 ubiquitin-protein ligase synoviolin [EC:6.3.2.19]
Length=595
Score = 37.0 bits (84), Expect = 0.016, Method: Composition-based stats.
Identities = 18/57 (31%), Positives = 31/57 (54%), Gaps = 0/57 (0%)
Query 2 YLFNDEFYTAVVKLASNKLALAIEYNFCLLMFLLFVKFILHLLVGRLRRLEVEQVLD 58
Y ++FY VV L + ++AI Y ++ L KF+ + G+LR E+E +L+
Sbjct 17 YYLKNQFYPTVVYLTKSSPSMAILYIQAFVLVFLLGKFMGKVFFGQLRAAEMEHLLE 73
> xla:431869 syvn1-a, MGC83718, hrd1; synovial apoptosis inhibitor
1, synoviolin; K10601 E3 ubiquitin-protein ligase synoviolin
[EC:6.3.2.19]
Length=605
Score = 36.2 bits (82), Expect = 0.025, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 31/57 (54%), Gaps = 0/57 (0%)
Query 2 YLFNDEFYTAVVKLASNKLALAIEYNFCLLMFLLFVKFILHLLVGRLRRLEVEQVLD 58
Y ++FY VV L + ++A+ Y ++ L KF+ + G+LR E+E +L+
Sbjct 17 YYLKNQFYPTVVYLTKSSPSMAVLYIQAFVLVFLLGKFMGKVFFGQLRAAEMEHLLE 73
> dre:335226 syvn1, wu:fk91f10, zgc:55735, zgc:77108; synovial
apoptosis inhibitor 1, synoviolin (EC:6.3.2.-); K10601 E3 ubiquitin-protein
ligase synoviolin [EC:6.3.2.19]
Length=625
Score = 33.1 bits (74), Expect = 0.22, Method: Composition-based stats.
Identities = 15/57 (26%), Positives = 29/57 (50%), Gaps = 0/57 (0%)
Query 2 YLFNDEFYTAVVKLASNKLALAIEYNFCLLMFLLFVKFILHLLVGRLRRLEVEQVLD 58
Y +FY VV L + ++A+ Y ++ L K + + G+LR E+E +++
Sbjct 23 YFLKHQFYPTVVYLTKSSPSMAVLYIQAFVLVFLLGKLMRKVFFGQLRAAEMEHLIE 79
> mmu:74126 Syvn1, 1200010C09Rik, AW211966, C85322, D530017H19Rik,
Hrd1; synovial apoptosis inhibitor 1, synoviolin; K10601
E3 ubiquitin-protein ligase synoviolin [EC:6.3.2.19]
Length=612
Score = 32.3 bits (72), Expect = 0.37, Method: Composition-based stats.
Identities = 16/57 (28%), Positives = 29/57 (50%), Gaps = 0/57 (0%)
Query 2 YLFNDEFYTAVVKLASNKLALAIEYNFCLLMFLLFVKFILHLLVGRLRRLEVEQVLD 58
Y +FY VV L + ++A+ Y ++ L K + + G+LR E+E +L+
Sbjct 23 YYLKHQFYPTVVYLTKSSPSMAVLYIQAFVLVFLLGKVMGKVFFGQLRAAEMEHLLE 79
> hsa:84447 SYVN1, HRD1, KIAA1810, MGC40372; synovial apoptosis
inhibitor 1, synoviolin (EC:6.3.2.-); K10601 E3 ubiquitin-protein
ligase synoviolin [EC:6.3.2.19]
Length=616
Score = 32.3 bits (72), Expect = 0.37, Method: Composition-based stats.
Identities = 16/57 (28%), Positives = 29/57 (50%), Gaps = 0/57 (0%)
Query 2 YLFNDEFYTAVVKLASNKLALAIEYNFCLLMFLLFVKFILHLLVGRLRRLEVEQVLD 58
Y +FY VV L + ++A+ Y ++ L K + + G+LR E+E +L+
Sbjct 23 YYLKHQFYPTVVYLTKSSPSMAVLYIQAFVLVFLLGKVMGKVFFGQLRAAEMEHLLE 79
> ath:AT1G08800 hypothetical protein
Length=1113
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 13/28 (46%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 21 ALAIEYNFCLLMFLLFVKFILHLLVGRL 48
ALA+ +N LLMF+LFV I ++ R
Sbjct 9 ALALAFNEWLLMFMLFVNSIFSYVIARF 36
Lambda K H
0.335 0.151 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2060478756
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40