bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2876_orf1
Length=113
Score E
Sequences producing significant alignments: (Bits) Value
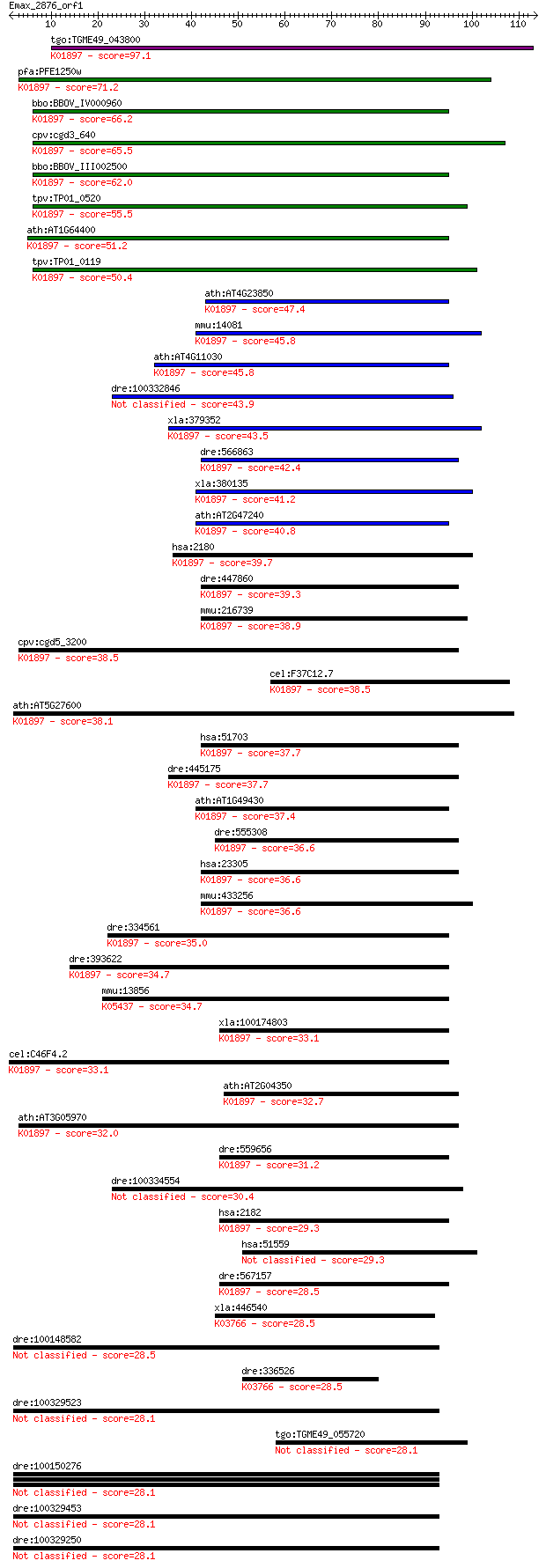
tgo:TGME49_043800 long-chain fatty acid CoA ligase, putative (... 97.1 1e-20
pfa:PFE1250w PfACS10; acyl-CoA synthetase, PfACS10 (EC:6.2.1.3... 71.2 7e-13
bbo:BBOV_IV000960 21.m02785; fatty acyl-CoA synthetase 3 (EC:6... 66.2 3e-11
cpv:cgd3_640 acyl-CoA synthetase ; K01897 long-chain acyl-CoA ... 65.5 4e-11
bbo:BBOV_III002500 17.m07241; fatty acyl-CoA synthetase family... 62.0 5e-10
tpv:TP01_0520 long-chain fatty acid CoA ligase; K01897 long-ch... 55.5 4e-08
ath:AT1G64400 long-chain-fatty-acid--CoA ligase, putative / lo... 51.2 8e-07
tpv:TP01_0119 long-chain fatty acid CoA ligase; K01897 long-ch... 50.4 1e-06
ath:AT4G23850 long-chain-fatty-acid--CoA ligase / long-chain a... 47.4 1e-05
mmu:14081 Acsl1, Acas, Acas1, Acs, FACS, Facl2; acyl-CoA synth... 45.8 3e-05
ath:AT4G11030 long-chain-fatty-acid--CoA ligase, putative / lo... 45.8 4e-05
dre:100332846 acyl-CoA synthetase long-chain family member 6-like 43.9 1e-04
xla:379352 MGC53832; similar to fatty acid Coenzyme A ligase; ... 43.5 2e-04
dre:566863 fa04h02, wu:fc05h07; wu:fa04h02; K01897 long-chain ... 42.4 4e-04
xla:380135 acsl1, MGC132122, MGC52902, facl2; acyl-CoA synthet... 41.2 9e-04
ath:AT2G47240 long-chain-fatty-acid--CoA ligase family protein... 40.8 0.001
hsa:2180 ACSL1, ACS1, FACL1, FACL2, LACS, LACS1, LACS2; acyl-C... 39.7 0.002
dre:447860 acsl5, zgc:92083; acyl-CoA synthetase long-chain fa... 39.3 0.003
mmu:216739 Acsl6, A330035H04Rik, AW050338, Facl6, LACS, Lacsl,... 38.9 0.004
cpv:cgd5_3200 acyl-CoA synthetase ; K01897 long-chain acyl-CoA... 38.5 0.005
cel:F37C12.7 acs-16; fatty Acid CoA Synthetase family member (... 38.5 0.006
ath:AT5G27600 LACS7; LACS7 (LONG-CHAIN ACYL-COA SYNTHETASE 7);... 38.1 0.008
hsa:51703 ACSL5, ACS2, ACS5, FACL5; acyl-CoA synthetase long-c... 37.7 0.009
dre:445175 zgc:101071 (EC:6.2.1.3); K01897 long-chain acyl-CoA... 37.7 0.010
ath:AT1G49430 LACS2; LACS2 (LONG-CHAIN ACYL-COA SYNTHETASE 2);... 37.4 0.012
dre:555308 acsl1, MGC110081, zgc:110081; acyl-CoA synthetase l... 36.6 0.020
hsa:23305 ACSL6, ACS2, FACL6, FLJ16173, KIAA0837, LACS_6, LACS... 36.6 0.020
mmu:433256 Acsl5, 1700030F05Rik, ACS2, ACS5, Facl5; acyl-CoA s... 36.6 0.022
dre:334561 acsl4l, fl49b07, si:dkeyp-20g2.4, wu:fl49b07; acyl-... 35.0 0.065
dre:393622 acsl4a, MGC66186, acsl4, zgc:66186; acyl-CoA synthe... 34.7 0.068
mmu:13856 Epo; erythropoietin; K05437 erythropoietin 34.7 0.073
xla:100174803 acsl4, acs4, acsl3, facl4, lacs4, mrx63, mrx68; ... 33.1 0.21
cel:C46F4.2 acs-17; fatty Acid CoA Synthetase family member (a... 33.1 0.24
ath:AT2G04350 long-chain-fatty-acid--CoA ligase family protein... 32.7 0.30
ath:AT3G05970 LACS6; LACS6 (long-chain acyl-CoA synthetase 6);... 32.0 0.45
dre:559656 acsl3b, acsl3, fb34c03, im:7155129, si:dkey-20f20.5... 31.2 0.96
dre:100334554 hypothetical protein LOC100334554 30.4 1.5
hsa:2182 ACSL4, ACS4, FACL4, LACS4, MRX63, MRX68; acyl-CoA syn... 29.3 3.0
hsa:51559 NT5DC3, FLJ11266, TU12B1-TY, TU12B1TY; 5'-nucleotida... 29.3 3.1
dre:567157 acsl3a, si:dkeyp-109h9.2; acyl-CoA synthetase long-... 28.5 5.0
xla:446540 b3gnt5-b, MGC80310, b3gn-t5, b3gnt5a, beta3gn-t5; U... 28.5 5.7
dre:100148582 protocadherin 2G30-like 28.5 5.8
dre:336526 b3gnt5a, Lc3, b3gnt5, fb12f12, wu:fb12f12, wu:fe50g... 28.5 6.1
dre:100329523 protocadherin 2G30-like 28.1 6.5
tgo:TGME49_055720 hypothetical protein 28.1 6.8
dre:100150276 protocadherin beta 16-like 28.1 7.3
dre:100329453 protocadherin 2 gamma 17-like 28.1 7.3
dre:100329250 protocadherin gamma subfamily A, 10-like 28.1 7.9
> tgo:TGME49_043800 long-chain fatty acid CoA ligase, putative
(EC:6.2.1.3); K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=748
Score = 97.1 bits (240), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 52/104 (50%), Positives = 66/104 (63%), Gaps = 4/104 (3%)
Query 10 VPSGESTADRTPPRRHCSSLDRLAENFSTNPVKNAWELFQMGVGKSAEGPCLGTRLVTPD 69
VP+G+ RT R S D + +NF+ P ++AW+LFQ GV S E CLGTR+ D
Sbjct 20 VPAGKK---RTSIYRSVFSPDAIVDNFADRPCQSAWDLFQRGVAISGEKRCLGTRVKNAD 76
Query 70 GRRGEYVWKSYREVERLALEAGSGLLN-GDFIPLLHFEQPGGQQ 112
G G Y WK+YREVE+LALE GSG+L+ D P LHFE+ QQ
Sbjct 77 GTLGPYQWKTYREVEQLALEVGSGILSLPDAAPKLHFEEETFQQ 120
> pfa:PFE1250w PfACS10; acyl-CoA synthetase, PfACS10 (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=673
Score = 71.2 bits (173), Expect = 7e-13, Method: Composition-based stats.
Identities = 38/101 (37%), Positives = 53/101 (52%), Gaps = 1/101 (0%)
Query 3 DVQYSVSVPSGESTADRTPPRRHCSSLDRLAENFSTNPVKNAWELFQMGVGKSAEGPCLG 62
++ YSV + S + T RH D+L ENF N WE+F K E C+G
Sbjct 7 EILYSVKI-SEPRDKNSTGIYRHPEYKDKLCENFDDKKFNNMWEVFDRVSTKFKERDCMG 65
Query 63 TRLVTPDGRRGEYVWKSYREVERLALEAGSGLLNGDFIPLL 103
R D +RG Y WK++ EV+ L ++ GSGLLN + PL+
Sbjct 66 VREKLEDNKRGAYQWKNFGEVKELIMKVGSGLLNMNACPLI 106
> bbo:BBOV_IV000960 21.m02785; fatty acyl-CoA synthetase 3 (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=681
Score = 66.2 bits (160), Expect = 3e-11, Method: Composition-based stats.
Identities = 33/89 (37%), Positives = 50/89 (56%), Gaps = 2/89 (2%)
Query 6 YSVSVPSGESTADRTPPRRHCSSLDRLAENFSTNPVKNAWELFQMGVGKSAEGPCLGTRL 65
Y+V VP T +P R+ +++D + N W++FQ G+ K+ + PC+G R
Sbjct 17 YAVPVPDTAQTG-YSPIYRN-AAVDTIKTALDYGNFHNMWDVFQHGLKKNPDAPCIGKRA 74
Query 66 VTPDGRRGEYVWKSYREVERLALEAGSGL 94
V PDG RG Y + +Y++VE AL GS L
Sbjct 75 VNPDGTRGAYSFMTYKQVETFALLIGSAL 103
> cpv:cgd3_640 acyl-CoA synthetase ; K01897 long-chain acyl-CoA
synthetase [EC:6.2.1.3]
Length=685
Score = 65.5 bits (158), Expect = 4e-11, Method: Composition-based stats.
Identities = 36/101 (35%), Positives = 50/101 (49%), Gaps = 0/101 (0%)
Query 6 YSVSVPSGESTADRTPPRRHCSSLDRLAENFSTNPVKNAWELFQMGVGKSAEGPCLGTRL 65
+SV + +G+ + +P R+ S N N WELF V K CLGTR
Sbjct 11 FSVELENGQVEPESSPIYRNPSYSKGELSNLQGLNANNLWELFVNSVNKYKNKKCLGTRK 70
Query 66 VTPDGRRGEYVWKSYREVERLALEAGSGLLNGDFIPLLHFE 106
+ DG GEYV+KSY E++ AL G +L D P+ +E
Sbjct 71 LNRDGTYGEYVFKSYEELKEEALNIGINILKMDLCPIRKYE 111
> bbo:BBOV_III002500 17.m07241; fatty acyl-CoA synthetase family
protein; K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=663
Score = 62.0 bits (149), Expect = 5e-10, Method: Composition-based stats.
Identities = 35/91 (38%), Positives = 48/91 (52%), Gaps = 7/91 (7%)
Query 6 YSVSVPSGESTADRTPPRRHCSSLDRLA--ENFSTNPVKNAWELFQMGVGKSAEGPCLGT 63
Y S SG S R P +D+L ++ + W++FQ G+ + + PCLG+
Sbjct 6 YPNSCGSGYSPIYRCP-----KYMDKLLSYKDVHDGKIATGWDIFQHGLSLNPDAPCLGS 60
Query 64 RLVTPDGRRGEYVWKSYREVERLALEAGSGL 94
R DG GEYV+KSY+EVE L GSGL
Sbjct 61 REKKADGSLGEYVFKSYKEVESLVQRFGSGL 91
> tpv:TP01_0520 long-chain fatty acid CoA ligase; K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=669
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 34/95 (35%), Positives = 49/95 (51%), Gaps = 3/95 (3%)
Query 6 YSVSVPSGESTADRTPPRRHCSSLDRL--AENFSTNPVKNAWELFQMGVGKSAEGPCLGT 63
YSV++ G TP R+ DRL E++ ++ W++F G+ S P LG+
Sbjct 5 YSVAL-EGTEEEGYTPVYRNPDYKDRLLSCEDYFDGKIQTGWDIFNRGLTLSRNKPFLGS 63
Query 64 RLVTPDGRRGEYVWKSYREVERLALEAGSGLLNGD 98
R+ DG GE+ + +Y EVE GSGLLN D
Sbjct 64 RVKNADGTFGEFKFMTYGEVEDQIKHFGSGLLNLD 98
> ath:AT1G64400 long-chain-fatty-acid--CoA ligase, putative /
long-chain acyl-CoA synthetase, putative; K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=665
Score = 51.2 bits (121), Expect = 8e-07, Method: Composition-based stats.
Identities = 29/95 (30%), Positives = 50/95 (52%), Gaps = 8/95 (8%)
Query 5 QYSVSVPSGESTADRTPPR-----RHCSSLDRLAENFSTNPVKNAWELFQMGVGKSAEGP 59
+Y V V G+ D P R + D E + + +AW++F++ V KS P
Sbjct 5 RYIVEVEKGKQGVDGGSPSVGPVYRSIYAKDGFPE--PPDDLVSAWDIFRLSVEKSPNNP 62
Query 60 CLGTRLVTPDGRRGEYVWKSYREVERLALEAGSGL 94
LG R + DG+ G+YVW++Y+EV + ++ G+ +
Sbjct 63 MLGRREIV-DGKAGKYVWQTYKEVHNVVIKLGNSI 96
> tpv:TP01_0119 long-chain fatty acid CoA ligase; K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=691
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 29/96 (30%), Positives = 49/96 (51%), Gaps = 4/96 (4%)
Query 6 YSVSVPSGESTADRTPPRRHCSSLDRL-AENFSTNPVKNAWELFQMGVGKSAEGPCLGTR 64
YSV +P E +P RH L A +F K +W+LFQ G+ ++ + C+G R
Sbjct 12 YSVPIPGTEEEG-FSPVYRHPKHEKILGASDFGD--FKTSWDLFQAGLRRNPDAECVGKR 68
Query 65 LVTPDGRRGEYVWKSYREVERLALEAGSGLLNGDFI 100
PDG+ G++ + +Y E + GS L++ + +
Sbjct 69 KRLPDGKLGDFEFNTYSEFFKTTKAVGSSLVHHNLV 104
> ath:AT4G23850 long-chain-fatty-acid--CoA ligase / long-chain
acyl-CoA synthetase; K01897 long-chain acyl-CoA synthetase
[EC:6.2.1.3]
Length=666
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 34/52 (65%), Gaps = 1/52 (1%)
Query 43 NAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSYREVERLALEAGSGL 94
+ W++F+M V K P LG R + DG+ G+YVW++Y+EV + ++ G+ L
Sbjct 46 SCWDVFRMSVEKYPNNPMLGRREIV-DGKPGKYVWQTYQEVYDIVMKLGNSL 96
> mmu:14081 Acsl1, Acas, Acas1, Acs, FACS, Facl2; acyl-CoA synthetase
long-chain family member 1 (EC:6.2.1.3); K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=699
Score = 45.8 bits (107), Expect = 3e-05, Method: Composition-based stats.
Identities = 26/61 (42%), Positives = 34/61 (55%), Gaps = 5/61 (8%)
Query 41 VKNAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSYREVERLALEAGSGLLNGDFI 100
V+ ++ FQ G+ S GPCLG+R P+ Y W SY+EV LA GSGL+ F
Sbjct 90 VRTMYDGFQRGIQVSNNGPCLGSR--KPNQ---PYEWISYKEVAELAECIGSGLIQKGFK 144
Query 101 P 101
P
Sbjct 145 P 145
> ath:AT4G11030 long-chain-fatty-acid--CoA ligase, putative /
long-chain acyl-CoA synthetase, putative (EC:6.2.1.3); K01897
long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=666
Score = 45.8 bits (107), Expect = 4e-05, Method: Composition-based stats.
Identities = 22/66 (33%), Positives = 40/66 (60%), Gaps = 4/66 (6%)
Query 32 LAENFSTNPV---KNAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSYREVERLAL 88
A+N NP+ ++ W++F+ V K LG R ++ +G+ G+YVWK+Y+EV + +
Sbjct 32 FAQNGFPNPIDGIQSCWDIFRTAVEKYPNNRMLGRREIS-NGKAGKYVWKTYKEVYDIVI 90
Query 89 EAGSGL 94
+ G+ L
Sbjct 91 KLGNSL 96
> dre:100332846 acyl-CoA synthetase long-chain family member 6-like
Length=1391
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 28/73 (38%), Positives = 35/73 (47%), Gaps = 5/73 (6%)
Query 23 RRHCSSLDRLAENFSTNPVKNAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSYRE 82
RR D +F K A+++FQ G+ + +GPCLG R R Y W SY E
Sbjct 785 RRSALLKDDTLLDFYYEDTKTAYDMFQRGLRIAGDGPCLGFR-----KPREPYQWISYTE 839
Query 83 VERLALEAGSGLL 95
V A GSGLL
Sbjct 840 VAERAQVLGSGLL 852
> xla:379352 MGC53832; similar to fatty acid Coenzyme A ligase;
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=698
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 27/67 (40%), Positives = 35/67 (52%), Gaps = 5/67 (7%)
Query 35 NFSTNPVKNAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSYREVERLALEAGSGL 94
+ + VK +++FQ GV S GPCLG R P+ Y W SY+EV A GS L
Sbjct 83 TYYYDDVKTIYDIFQRGVRVSNNGPCLGHR--KPN---QPYEWISYKEVAERAEFVGSAL 137
Query 95 LNGDFIP 101
L+ F P
Sbjct 138 LHRGFKP 144
> dre:566863 fa04h02, wu:fc05h07; wu:fa04h02; K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=633
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 25/55 (45%), Positives = 33/55 (60%), Gaps = 5/55 (9%)
Query 42 KNAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSYREVERLALEAGSGLLN 96
K +E+FQ G+ + +GP LG+RL P+ Y W SYREV A GSGLL+
Sbjct 25 KTMYEVFQRGLHITGDGPFLGSRL--PNQ---PYTWLSYREVSTRAEYVGSGLLS 74
> xla:380135 acsl1, MGC132122, MGC52902, facl2; acyl-CoA synthetase
long-chain family member 1 protein (EC:6.2.1.3); K01897
long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=698
Score = 41.2 bits (95), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 25/59 (42%), Positives = 32/59 (54%), Gaps = 5/59 (8%)
Query 41 VKNAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSYREVERLALEAGSGLLNGDF 99
VK +++FQ G+ S GPCLG R P+ Y W SY+EV A GS LL+ F
Sbjct 89 VKTIYDIFQRGMRVSNNGPCLGHR--KPN---QPYEWISYKEVSERAEYVGSALLHRGF 142
> ath:AT2G47240 long-chain-fatty-acid--CoA ligase family protein
/ long-chain acyl-CoA synthetase family protein (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=660
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 30/54 (55%), Gaps = 1/54 (1%)
Query 41 VKNAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSYREVERLALEAGSGL 94
+ AW++F V K + LG R + D + G Y+WK+Y+EV L+ GS L
Sbjct 41 ITTAWDIFSKSVEKFPDNNMLGWRRIV-DEKVGPYMWKTYKEVYEEVLQIGSAL 93
> hsa:2180 ACSL1, ACS1, FACL1, FACL2, LACS, LACS1, LACS2; acyl-CoA
synthetase long-chain family member 1 (EC:6.2.1.3); K01897
long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=698
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 33/64 (51%), Gaps = 5/64 (7%)
Query 36 FSTNPVKNAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSYREVERLALEAGSGLL 95
+ + V +E FQ G+ S GPCLG+R PD Y W SY++V L+ GS L+
Sbjct 84 YFYDDVTTLYEGFQRGIQVSNNGPCLGSR--KPD---QPYEWLSYKQVAELSECIGSALI 138
Query 96 NGDF 99
F
Sbjct 139 QKGF 142
> dre:447860 acsl5, zgc:92083; acyl-CoA synthetase long-chain
family member 5 (EC:6.2.1.3); K01897 long-chain acyl-CoA synthetase
[EC:6.2.1.3]
Length=681
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 22/56 (39%), Positives = 32/56 (57%), Gaps = 7/56 (12%)
Query 42 KNAWELFQMGVGKSAEGPCLGTRLVTPDGRRGE-YVWKSYREVERLALEAGSGLLN 96
K +E+FQ G+ S GPCLG R ++G+ Y W Y++V A GSGL++
Sbjct 75 KTLYEVFQRGLRVSGNGPCLGFR------KKGQPYQWLKYKQVSDRAEFLGSGLIH 124
> mmu:216739 Acsl6, A330035H04Rik, AW050338, Facl6, LACS, Lacsl,
MGC36650, mKIAA0837; acyl-CoA synthetase long-chain family
member 6 (EC:6.2.1.3); K01897 long-chain acyl-CoA synthetase
[EC:6.2.1.3]
Length=722
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 23/57 (40%), Positives = 32/57 (56%), Gaps = 5/57 (8%)
Query 42 KNAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSYREVERLALEAGSGLLNGD 98
+ +++F+ G+ S GPCLG R P+ Y W SY+EV + A GSGLL D
Sbjct 115 RTMYQVFRRGLSISGNGPCLGFR--KPEQ---PYQWLSYQEVAKRAEFLGSGLLQHD 166
> cpv:cgd5_3200 acyl-CoA synthetase ; K01897 long-chain acyl-CoA
synthetase [EC:6.2.1.3]
Length=683
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 29/95 (30%), Positives = 44/95 (46%), Gaps = 3/95 (3%)
Query 3 DVQYSVSVPSGESTADRTPPRRHCSSLD-RLAENFSTNPVKNAWELFQMGVGKSAEGPCL 61
D YS + G ++ T RR +++ L NF ++N W + G S G +
Sbjct 14 DFIYSCPI-EGTGDSNSTEIRRCKETINSELTSNFHEK-LENCWGILLHGKEASNNGDYM 71
Query 62 GTRLVTPDGRRGEYVWKSYREVERLALEAGSGLLN 96
G R+ +G +Y + Y V R A E GSGLL+
Sbjct 72 GVRVPLKNGELSDYKFVKYSYVIRKAKEVGSGLLH 106
> cel:F37C12.7 acs-16; fatty Acid CoA Synthetase family member
(acs-16); K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=731
Score = 38.5 bits (88), Expect = 0.006, Method: Composition-based stats.
Identities = 25/64 (39%), Positives = 31/64 (48%), Gaps = 18/64 (28%)
Query 57 EGPCLGTRLV-------TPDGR------RGEYVWKSYREVERLALEAGSGLLNGDFIPLL 103
E PCLGTR + P GR GEYVW+SY+EVE +G+ + L
Sbjct 108 ESPCLGTRQLIEVHEDKQPGGRVFEKWHLGEYVWQSYKEVETQVARLAAGIKD-----LA 162
Query 104 HFEQ 107
H EQ
Sbjct 163 HEEQ 166
> ath:AT5G27600 LACS7; LACS7 (LONG-CHAIN ACYL-COA SYNTHETASE 7);
long-chain-fatty-acid-CoA ligase/ protein binding (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=700
Score = 38.1 bits (87), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 37/113 (32%), Positives = 47/113 (41%), Gaps = 8/113 (7%)
Query 2 KDVQYSVSVPSGESTADRTPPRRHCSSLDRLAENFSTNP-VKNAWELFQMGVGKSAEGPC 60
K+ YSV +P T + R S +L F +P + + F V AE
Sbjct 44 KEDSYSVVLPEKLDTG-KWNVYRSKRSPTKLVSRFPDHPEIGTLHDNFVHAVETYAENKY 102
Query 61 LGTRLVTPDGRRGEYVWKSYREVERLALEAGSGLL-----NGDFIPLLHFEQP 108
LGTR V DG GEY W +Y E GSGLL GD + L +P
Sbjct 103 LGTR-VRSDGTIGEYSWMTYGEAASERQAIGSGLLFHGVNQGDCVGLYFINRP 154
> hsa:51703 ACSL5, ACS2, ACS5, FACL5; acyl-CoA synthetase long-chain
family member 5 (EC:6.2.1.3); K01897 long-chain acyl-CoA
synthetase [EC:6.2.1.3]
Length=739
Score = 37.7 bits (86), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 23/55 (41%), Positives = 30/55 (54%), Gaps = 5/55 (9%)
Query 42 KNAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSYREVERLALEAGSGLLN 96
K +E+FQ G+ S GPCLG R P+ Y W SY++V A GS LL+
Sbjct 131 KTMYEVFQRGLAVSDNGPCLGYR--KPN---QPYRWLSYKQVSDRAEYLGSCLLH 180
> dre:445175 zgc:101071 (EC:6.2.1.3); K01897 long-chain acyl-CoA
synthetase [EC:6.2.1.3]
Length=697
Score = 37.7 bits (86), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 29/62 (46%), Gaps = 5/62 (8%)
Query 35 NFSTNPVKNAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSYREVERLALEAGSGL 94
+F K +E F G+ +S GPCLG+R + Y W SY EV A GS
Sbjct 83 SFYYEDAKTMYECFHRGLRESKNGPCLGSR-----KPKQPYEWLSYSEVIERAEHLGSAF 137
Query 95 LN 96
L+
Sbjct 138 LH 139
> ath:AT1G49430 LACS2; LACS2 (LONG-CHAIN ACYL-COA SYNTHETASE 2);
long-chain-fatty-acid-CoA ligase (EC:6.2.1.3); K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=665
Score = 37.4 bits (85), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 26/54 (48%), Gaps = 1/54 (1%)
Query 41 VKNAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSYREVERLALEAGSGL 94
+ + W+ F V K LG R VT D + G Y W +Y+E A+ GS +
Sbjct 44 IDSPWQFFSEAVKKYPNEQMLGQR-VTTDSKVGPYTWITYKEAHDAAIRIGSAI 96
> dre:555308 acsl1, MGC110081, zgc:110081; acyl-CoA synthetase
long-chain family member 1 (EC:6.2.1.3); K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=697
Score = 36.6 bits (83), Expect = 0.020, Method: Composition-based stats.
Identities = 22/53 (41%), Positives = 29/53 (54%), Gaps = 7/53 (13%)
Query 45 WELFQMGVGKSAEGPCLGTRLVTPDGRRGE-YVWKSYREVERLALEAGSGLLN 96
+E F G+ GPCLG+R + G+ Y W SY+EV A AGS LL+
Sbjct 93 YEFFLRGLRVPNNGPCLGSR------KAGQPYKWLSYKEVADRAEFAGSALLH 139
> hsa:23305 ACSL6, ACS2, FACL6, FLJ16173, KIAA0837, LACS_6, LACS2,
LACS5; acyl-CoA synthetase long-chain family member 6 (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=722
Score = 36.6 bits (83), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 21/55 (38%), Positives = 29/55 (52%), Gaps = 5/55 (9%)
Query 42 KNAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSYREVERLALEAGSGLLN 96
+ +++F+ G+ S GPCLG R + Y W SY+EV A GSGLL
Sbjct 115 RTMYQVFRRGLSISGNGPCLGFR-----KPKQPYQWLSYQEVADRAEFLGSGLLQ 164
> mmu:433256 Acsl5, 1700030F05Rik, ACS2, ACS5, Facl5; acyl-CoA
synthetase long-chain family member 5 (EC:6.2.1.3); K01897
long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=683
Score = 36.6 bits (83), Expect = 0.022, Method: Composition-based stats.
Identities = 23/58 (39%), Positives = 30/58 (51%), Gaps = 5/58 (8%)
Query 42 KNAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSYREVERLALEAGSGLLNGDF 99
K +E FQ G+ S GPCLG R P+ Y W SY++V A GS LL+ +
Sbjct 75 KTLYENFQRGLAVSDNGPCLGYR--KPN---QPYKWISYKQVSDRAEYLGSCLLHKGY 127
> dre:334561 acsl4l, fl49b07, si:dkeyp-20g2.4, wu:fl49b07; acyl-CoA
synthetase long-chain family member 4, like; K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=671
Score = 35.0 bits (79), Expect = 0.065, Method: Compositional matrix adjust.
Identities = 28/86 (32%), Positives = 35/86 (40%), Gaps = 14/86 (16%)
Query 22 PRRHCSSLDRLAENFSTNPVKNAWELFQMGVGKSAEGPCLGTRLV-------TPDGR--- 71
P R S+D L V +LF V + PCLGTR V P+G+
Sbjct 18 PYRAVESIDALVTQ-GFEGVDTLDKLFVHAVKSFNQAPCLGTREVLSEEEEKQPNGKIFK 76
Query 72 ---RGEYVWKSYREVERLALEAGSGL 94
G Y W SY V++ GSGL
Sbjct 77 KLVLGNYSWMSYEAVDQAVESFGSGL 102
> dre:393622 acsl4a, MGC66186, acsl4, zgc:66186; acyl-CoA synthetase
long-chain family member 4a (EC:6.2.1.3); K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=706
Score = 34.7 bits (78), Expect = 0.068, Method: Composition-based stats.
Identities = 32/96 (33%), Positives = 45/96 (46%), Gaps = 17/96 (17%)
Query 14 ESTADRTPPRRHCSSLDRLAENFSTN-PVKNAWE-LFQMGVGKSAEGPCLGTRLV----- 66
+ST+D+ P S DR + + P K+ + LF V + + CLGTR V
Sbjct 45 KSTSDQ--PDGPYRSTDRFESLVTVDFPGKDTLDKLFDYAVERFGKLDCLGTREVLSGDN 102
Query 67 --TPDGR------RGEYVWKSYREVERLALEAGSGL 94
P+G+ GEY WKSY E++ GSGL
Sbjct 103 ETQPNGKVFKKLILGEYKWKSYEELDIEVSHFGSGL 138
> mmu:13856 Epo; erythropoietin; K05437 erythropoietin
Length=192
Score = 34.7 bits (78), Expect = 0.073, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 35/74 (47%), Gaps = 8/74 (10%)
Query 21 PPRRHCSSLDRLAENFSTNPVKNAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSY 80
PPR C S R+ E + + A E + +G AEGP L + PD + Y WK
Sbjct 28 PPRLICDS--RVLERY----ILEAKEAENVTMG-CAEGPRLSENITVPDTKVNFYAWKRM 80
Query 81 REVERLALEAGSGL 94
EVE A+E GL
Sbjct 81 -EVEEQAIEVWQGL 93
> xla:100174803 acsl4, acs4, acsl3, facl4, lacs4, mrx63, mrx68;
acyl-CoA synthetase long-chain family member 4 (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=670
Score = 33.1 bits (74), Expect = 0.21, Method: Composition-based stats.
Identities = 22/62 (35%), Positives = 28/62 (45%), Gaps = 13/62 (20%)
Query 46 ELFQMGVGKSAEGPCLGTRLV-------TPDGR------RGEYVWKSYREVERLALEAGS 92
+LF+ GV + CL TR + P+G+ GEY W SY EV GS
Sbjct 40 KLFEHGVATFGKKDCLSTREILSEENEKQPNGKVFKKLILGEYKWLSYEEVSLRVHHLGS 99
Query 93 GL 94
GL
Sbjct 100 GL 101
> cel:C46F4.2 acs-17; fatty Acid CoA Synthetase family member
(acs-17); K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=718
Score = 33.1 bits (74), Expect = 0.24, Method: Composition-based stats.
Identities = 28/107 (26%), Positives = 48/107 (44%), Gaps = 14/107 (13%)
Query 1 AKDVQYSVSVPSGESTADRTPPRRHCSSLDRLAENFSTNPVKNAWELFQMGVGKSAEGPC 60
A+ ++ S V + D T P ++ S+ D L N S + + ++ V + PC
Sbjct 49 AEKLEKSERVKAEPDNGDPTTPWKNVSAKDGLM-NSSFEGIHDLGTQWEECVRRYGNQPC 107
Query 61 LGTRLV-------TPDGR------RGEYVWKSYREVERLALEAGSGL 94
+GTR V +G+ G+Y W+SY +V++ SGL
Sbjct 108 MGTREVLKVHKEKQSNGKIFEKWQMGDYHWRSYAQVDKRVNMIASGL 154
> ath:AT2G04350 long-chain-fatty-acid--CoA ligase family protein
/ long-chain acyl-CoA synthetase family protein (LACS8) (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=720
Score = 32.7 bits (73), Expect = 0.30, Method: Composition-based stats.
Identities = 22/63 (34%), Positives = 29/63 (46%), Gaps = 13/63 (20%)
Query 47 LFQMGVGKSAEGPCLGTR-------LVTPDGRR------GEYVWKSYREVERLALEAGSG 93
LF+ K ++ LGTR + DGR+ GEY W+SY EV SG
Sbjct 86 LFEQSCKKYSKDRLLGTREFIDKEFITASDGRKFEKLHLGEYKWQSYGEVFERVCNFASG 145
Query 94 LLN 96
L+N
Sbjct 146 LVN 148
> ath:AT3G05970 LACS6; LACS6 (long-chain acyl-CoA synthetase 6);
long-chain-fatty-acid-CoA ligase (EC:6.2.1.3); K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=701
Score = 32.0 bits (71), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 29/95 (30%), Positives = 41/95 (43%), Gaps = 3/95 (3%)
Query 3 DVQYSVSVPSGESTADRTPPRRHCSSLDRLAENFSTNP-VKNAWELFQMGVGKSAEGPCL 61
D YSV +P +T R S +L F +P + + F+ V + L
Sbjct 45 DNGYSVVLPEKLNTGSWNVYRSAKSPF-KLVSRFPDHPDIATLHDNFEHAVHDFRDYKYL 103
Query 62 GTRLVTPDGRRGEYVWKSYREVERLALEAGSGLLN 96
GTR V DG G+Y W +Y E GSGL++
Sbjct 104 GTR-VRVDGTVGDYKWMTYGEAGTARTALGSGLVH 137
> dre:559656 acsl3b, acsl3, fb34c03, im:7155129, si:dkey-20f20.5,
wu:fa07a08, wu:fb34c03; acyl-CoA synthetase long-chain family
member 3b; K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=711
Score = 31.2 bits (69), Expect = 0.96, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 30/62 (48%), Gaps = 13/62 (20%)
Query 46 ELFQMGVGKSAEGPCLGTRL-------VTPDGR------RGEYVWKSYREVERLALEAGS 92
++F+ V E CLGTR + P+G+ G+Y W SY++ LA GS
Sbjct 81 KVFEYAVVHFPERDCLGTRELLSEEDEIQPNGKVFKKVILGDYNWLSYQDTFHLAQRFGS 140
Query 93 GL 94
GL
Sbjct 141 GL 142
> dre:100334554 hypothetical protein LOC100334554
Length=328
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 19/75 (25%), Positives = 35/75 (46%), Gaps = 3/75 (4%)
Query 23 RRHCSSLDRLAENFSTNPVKNAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSYRE 82
R+HC+S + + W+ F + G ++ CL R V GR+ EY +
Sbjct 242 RKHCNSKCCEMQKRTIAKSSPVWKYFSLKEGDCSKAVCLMCRAVISRGRK-EYTTSAL-- 298
Query 83 VERLALEAGSGLLNG 97
++ L ++ G+ L+ G
Sbjct 299 LKHLRMKHGNALIFG 313
> hsa:2182 ACSL4, ACS4, FACL4, LACS4, MRX63, MRX68; acyl-CoA synthetase
long-chain family member 4 (EC:6.2.1.3); K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=670
Score = 29.3 bits (64), Expect = 3.0, Method: Composition-based stats.
Identities = 21/62 (33%), Positives = 27/62 (43%), Gaps = 13/62 (20%)
Query 46 ELFQMGVGKSAEGPCLGTRLV-------TPDGR------RGEYVWKSYREVERLALEAGS 92
+LF V K + LGTR + P+G+ G Y W +Y EV R GS
Sbjct 40 KLFDHAVSKFGKKDSLGTREILSEENEMQPNGKVFKKLILGNYKWMNYLEVNRRVNNFGS 99
Query 93 GL 94
GL
Sbjct 100 GL 101
> hsa:51559 NT5DC3, FLJ11266, TU12B1-TY, TU12B1TY; 5'-nucleotidase
domain containing 3
Length=548
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 23/62 (37%), Positives = 28/62 (45%), Gaps = 14/62 (22%)
Query 51 GVGKSAEG-PCLGTRL-------VTPDGRRGEYVWKSYREVER----LALEAGSGLLNGD 98
G G +A G PC G PD +R Y+W+ YRE +R L S LLN D
Sbjct 26 GCGTAARGRPCAGPARPLCTAPGTAPDMKR--YLWERYREAKRSTEELVPSIMSNLLNPD 83
Query 99 FI 100
I
Sbjct 84 AI 85
> dre:567157 acsl3a, si:dkeyp-109h9.2; acyl-CoA synthetase long-chain
family member 3a (EC:6.2.1.3); K01897 long-chain acyl-CoA
synthetase [EC:6.2.1.3]
Length=713
Score = 28.5 bits (62), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 27/62 (43%), Gaps = 13/62 (20%)
Query 46 ELFQMGVGKSAEGPCLGTRLVTP-------DGR------RGEYVWKSYREVERLALEAGS 92
++FQ V CLGTR + DG+ GEY W SY +V + GS
Sbjct 83 QVFQSSVSHFPNSDCLGTRELLSEEDETQEDGKVFKKVILGEYRWMSYEQVFKEVNAFGS 142
Query 93 GL 94
GL
Sbjct 143 GL 144
> xla:446540 b3gnt5-b, MGC80310, b3gn-t5, b3gnt5a, beta3gn-t5;
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase
5 (EC:2.4.1.-); K03766 beta-1,3-N-acetylglucosaminyltransferase
5 [EC:2.4.1.206]
Length=377
Score = 28.5 bits (62), Expect = 5.7, Method: Composition-based stats.
Identities = 13/49 (26%), Positives = 26/49 (53%), Gaps = 2/49 (4%)
Query 45 WELFQMGVGKSAEGPCLGTRLVTPDGRRG--EYVWKSYREVERLALEAG 91
+ ++ G GKS PC+ +++T G G +Y+W+ + +L +G
Sbjct 298 YHVYFSGEGKSPYHPCIYNKMMTSHGHLGDLDYLWRQATDSNVKSLSSG 346
> dre:100148582 protocadherin 2G30-like
Length=800
Score = 28.5 bits (62), Expect = 5.8, Method: Composition-based stats.
Identities = 26/100 (26%), Positives = 44/100 (44%), Gaps = 14/100 (14%)
Query 2 KDVQYSVSVPSGESTADRTPPRRHCSSLDRLAENFSTN------PVKNAWELFQMGVGKS 55
K+ Q+ V G S PP +S+ L ++ + N PV++ + V +S
Sbjct 516 KEFQFRVKAQDGGS-----PPLSSNASVKILIQDQNDNAPQVLYPVQSGASVVAEIVPRS 570
Query 56 AEGPCLGTRLVTPDGRRGEYVWKSY---REVERLALEAGS 92
A+ L T++V D G+ W SY + +R E G+
Sbjct 571 ADVGYLVTKVVAVDVDSGQNAWLSYKLHKATDRALFEVGA 610
> dre:336526 b3gnt5a, Lc3, b3gnt5, fb12f12, wu:fb12f12, wu:fe50g08;
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase
5a (EC:2.4.1.-); K03766 beta-1,3-N-acetylglucosaminyltransferase
5 [EC:2.4.1.206]
Length=379
Score = 28.5 bits (62), Expect = 6.1, Method: Composition-based stats.
Identities = 11/31 (35%), Positives = 19/31 (61%), Gaps = 2/31 (6%)
Query 51 GVGKSAEGPCLGTRLVTPDGRRGE--YVWKS 79
G GK+ PC+ +++T G G+ Y+WK+
Sbjct 306 GEGKTPYHPCIYEKMITSHGHEGDIRYLWKA 336
> dre:100329523 protocadherin 2G30-like
Length=800
Score = 28.1 bits (61), Expect = 6.5, Method: Composition-based stats.
Identities = 26/100 (26%), Positives = 44/100 (44%), Gaps = 14/100 (14%)
Query 2 KDVQYSVSVPSGESTADRTPPRRHCSSLDRLAENFSTN------PVKNAWELFQMGVGKS 55
K+ Q+ V G S PP +S+ L ++ + N PV++ + V +S
Sbjct 516 KEFQFRVKAQDGGS-----PPLSSNASVKILIQDQNDNAPQVLYPVQSGASVVAEIVPRS 570
Query 56 AEGPCLGTRLVTPDGRRGEYVWKSY---REVERLALEAGS 92
A+ L T++V D G+ W SY + +R E G+
Sbjct 571 ADVGYLVTKVVAVDVDSGQNAWLSYKLHKATDRALFEVGA 610
> tgo:TGME49_055720 hypothetical protein
Length=458
Score = 28.1 bits (61), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 58 GPCLGTRLVTPDGRRGEYVWKSYREVERLALEAGSGLLNGD 98
G C G + PDGRR ++K RE+ L+ LL G+
Sbjct 162 GGCDGQHRLGPDGRRKTNIFKLEREIFLLSASQSRVLLGGE 202
> dre:100150276 protocadherin beta 16-like
Length=3331
Score = 28.1 bits (61), Expect = 7.3, Method: Composition-based stats.
Identities = 26/100 (26%), Positives = 44/100 (44%), Gaps = 14/100 (14%)
Query 2 KDVQYSVSVPSGESTADRTPPRRHCSSLDRLAENFSTN------PVKNAWELFQMGVGKS 55
K+ Q+ V G S PP +S+ L ++ + N PV++ + V +S
Sbjct 1151 KEFQFRVKAQDGGS-----PPLSSNASVKILIQDQNDNAPQVLYPVQSGASVVAEIVPRS 1205
Query 56 AEGPCLGTRLVTPDGRRGEYVWKSY---REVERLALEAGS 92
A+ L T++V D G+ W SY + +R E G+
Sbjct 1206 ADVGYLVTKVVAVDVDSGQNAWLSYKLHKATDRALFEVGA 1245
Score = 28.1 bits (61), Expect = 7.3, Method: Composition-based stats.
Identities = 26/100 (26%), Positives = 44/100 (44%), Gaps = 14/100 (14%)
Query 2 KDVQYSVSVPSGESTADRTPPRRHCSSLDRLAENFSTN------PVKNAWELFQMGVGKS 55
K+ Q+ V G S PP +S+ L ++ + N PV++ + V +S
Sbjct 1926 KEFQFRVKAQDGGS-----PPLSSNASVKILIQDQNDNAPQVLYPVQSGASVVAEIVPRS 1980
Query 56 AEGPCLGTRLVTPDGRRGEYVWKSY---REVERLALEAGS 92
A+ L T++V D G+ W SY + +R E G+
Sbjct 1981 ADVGYLVTKVVAVDVDSGQNAWLSYKLHKATDRALFEVGA 2020
Score = 28.1 bits (61), Expect = 7.3, Method: Composition-based stats.
Identities = 26/100 (26%), Positives = 44/100 (44%), Gaps = 14/100 (14%)
Query 2 KDVQYSVSVPSGESTADRTPPRRHCSSLDRLAENFSTN------PVKNAWELFQMGVGKS 55
K+ Q+ V G S PP +S+ L ++ + N PV++ + V +S
Sbjct 3045 KEFQFRVKAQDGGS-----PPLSSNASVKILIQDQNDNAPQVLYPVQSGASVVAEIVPRS 3099
Query 56 AEGPCLGTRLVTPDGRRGEYVWKSY---REVERLALEAGS 92
A+ L T++V D G+ W SY + +R E G+
Sbjct 3100 ADVGYLVTKVVAVDVDSGQNAWLSYKLHKATDRALFEVGA 3139
> dre:100329453 protocadherin 2 gamma 17-like
Length=800
Score = 28.1 bits (61), Expect = 7.3, Method: Composition-based stats.
Identities = 26/100 (26%), Positives = 44/100 (44%), Gaps = 14/100 (14%)
Query 2 KDVQYSVSVPSGESTADRTPPRRHCSSLDRLAENFSTN------PVKNAWELFQMGVGKS 55
K+ Q+ V G S PP +S+ L ++ + N PV++ + V +S
Sbjct 516 KEFQFRVKAQDGGS-----PPLSSNASVKILIQDQNDNAPQVLYPVQSGASVVAEIVPRS 570
Query 56 AEGPCLGTRLVTPDGRRGEYVWKSY---REVERLALEAGS 92
A+ L T++V D G+ W SY + +R E G+
Sbjct 571 ADVGYLVTKVVAVDVDSGQNAWLSYKLHKATDRALFEVGA 610
> dre:100329250 protocadherin gamma subfamily A, 10-like
Length=1572
Score = 28.1 bits (61), Expect = 7.9, Method: Composition-based stats.
Identities = 26/100 (26%), Positives = 44/100 (44%), Gaps = 14/100 (14%)
Query 2 KDVQYSVSVPSGESTADRTPPRRHCSSLDRLAENFSTN------PVKNAWELFQMGVGKS 55
K+ Q+ V G S PP +S+ L ++ + N PV++ + V +S
Sbjct 1288 KEFQFRVKAQDGGS-----PPLSSNASVKILIQDQNDNAPQVLYPVQSGASVVAEIVPRS 1342
Query 56 AEGPCLGTRLVTPDGRRGEYVWKSY---REVERLALEAGS 92
A+ L T++V D G+ W SY + +R E G+
Sbjct 1343 ADVGYLVTKVVAVDVDSGQNAWLSYKLHKATDRALFEVGA 1382
Lambda K H
0.315 0.135 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2052546600
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40