bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2858_orf1
Length=93
Score E
Sequences producing significant alignments: (Bits) Value
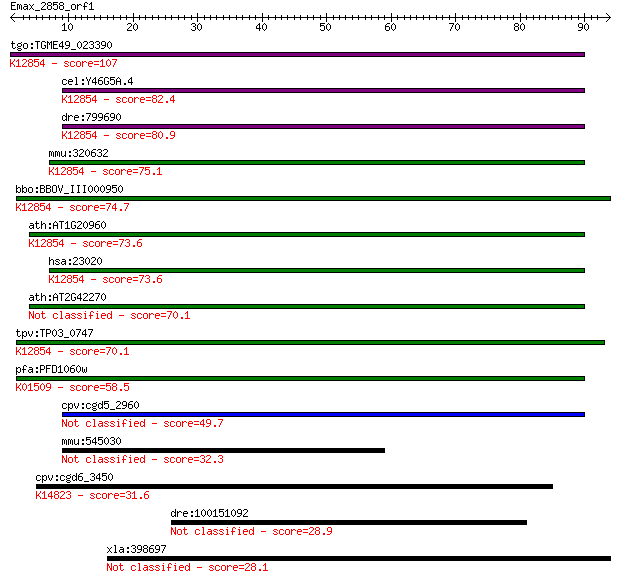
tgo:TGME49_023390 sec63 domain-containing DEAD/DEAH box helica... 107 1e-23
cel:Y46G5A.4 hypothetical protein; K12854 pre-mRNA-splicing he... 82.4 3e-16
dre:799690 fb63a09; wu:fb63a09 (EC:3.6.4.13); K12854 pre-mRNA-... 80.9 9e-16
mmu:320632 Snrnp200, A330064G03Rik, Ascc3l1, BC011390, HELIC2,... 75.1 5e-14
bbo:BBOV_III000950 17.m07111; sec63 domain containing protein ... 74.7 7e-14
ath:AT1G20960 emb1507 (embryo defective 1507); ATP binding / A... 73.6 1e-13
hsa:23020 SNRNP200, ASCC3L1, BRR2, FLJ11521, HELIC2, RP33, U5-... 73.6 1e-13
ath:AT2G42270 U5 small nuclear ribonucleoprotein helicase, put... 70.1 2e-12
tpv:TP03_0747 ATP-dependent RNA helicase; K12854 pre-mRNA-spli... 70.1 2e-12
pfa:PFD1060w U5 small nuclear ribonucleoprotein-specific prote... 58.5 5e-09
cpv:cgd5_2960 U5snrp Brr2 SFII RNA helicase (sec63 and the sec... 49.7 2e-06
mmu:545030 Wdfy4; WD repeat and FYVE domain containing 4 32.3
cpv:cgd6_3450 EBNA1 binding protein 2; nucleolar protein p40 ;... 31.6 0.70
dre:100151092 efcab6; EF-hand calcium binding domain 6 28.9
xla:398697 mtus1, fbx27; microtubule associated tumor suppress... 28.1 6.9
> tgo:TGME49_023390 sec63 domain-containing DEAD/DEAH box helicase,
putative ; K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2198
Score = 107 bits (266), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 60/89 (67%), Positives = 73/89 (82%), Gaps = 0/89 (0%)
Query 1 ADVTENFFYRPTTRVTNQVYEQLLVVLQQQLGDRPDEVLRGAADEVLQVLKEDGLKEGER 60
ADVTE FFY+PTT+ T VYEQLLV LQQQLGD+PDEVL+GAADEVL LK DG +E E+
Sbjct 93 ADVTEIFFYKPTTQQTRLVYEQLLVTLQQQLGDQPDEVLKGAADEVLAALKADGCRESEK 152
Query 61 KRGVEGVLGPITQERFTKLFQLSKSITDF 89
K+ VE VLGP+ + FT+L QL+K+ITD+
Sbjct 153 KKNVEQVLGPVNSDVFTRLHQLAKNITDY 181
> cel:Y46G5A.4 hypothetical protein; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2145
Score = 82.4 bits (202), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 41/81 (50%), Positives = 53/81 (65%), Gaps = 0/81 (0%)
Query 9 YRPTTRVTNQVYEQLLVVLQQQLGDRPDEVLRGAADEVLQVLKEDGLKEGERKRGVEGVL 68
Y+P T+ T Q YE +L + LGD P EVL GAADEVL LK D ++ E+K+ VE +L
Sbjct 97 YKPRTQETKQTYEVILSFILDALGDVPREVLCGAADEVLLTLKNDKFRDKEKKKEVEALL 156
Query 69 GPITQERFTKLFQLSKSITDF 89
GP+T +R L LSK I+DF
Sbjct 157 GPLTDDRIAVLINLSKKISDF 177
> dre:799690 fb63a09; wu:fb63a09 (EC:3.6.4.13); K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2134
Score = 80.9 bits (198), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 39/81 (48%), Positives = 54/81 (66%), Gaps = 0/81 (0%)
Query 9 YRPTTRVTNQVYEQLLVVLQQQLGDRPDEVLRGAADEVLQVLKEDGLKEGERKRGVEGVL 68
Y+P T+ T + YE LL +Q LGD+P ++L GAADEVL VLK D +++ ER+R VE +L
Sbjct 102 YKPKTKETRETYEILLSFIQAALGDQPRDILCGAADEVLAVLKNDKMRDKERRREVEQLL 161
Query 69 GPITQERFTKLFQLSKSITDF 89
GP R+ L L K I+D+
Sbjct 162 GPTDDTRYHVLVNLGKKISDY 182
> mmu:320632 Snrnp200, A330064G03Rik, Ascc3l1, BC011390, HELIC2,
KIAA0788, U5-200-KD, U5-200KD; small nuclear ribonucleoprotein
200 (U5) (EC:3.6.4.13); K12854 pre-mRNA-splicing helicase
BRR2 [EC:3.6.4.13]
Length=2136
Score = 75.1 bits (183), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 37/83 (44%), Positives = 53/83 (63%), Gaps = 0/83 (0%)
Query 7 FFYRPTTRVTNQVYEQLLVVLQQQLGDRPDEVLRGAADEVLQVLKEDGLKEGERKRGVEG 66
Y+P T+ T + YE LL +Q LGD+P ++L GAADEVL VLK + L++ ER+R ++
Sbjct 100 IIYKPKTKETRETYEVLLSFIQAALGDQPRDILCGAADEVLAVLKNEKLRDKERRREIDL 159
Query 67 VLGPITQERFTKLFQLSKSITDF 89
+LG R+ L L K ITD+
Sbjct 160 LLGQTDDTRYHVLVNLGKKITDY 182
> bbo:BBOV_III000950 17.m07111; sec63 domain containing protein
(EC:3.6.1.-); K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2133
Score = 74.7 bits (182), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 36/92 (39%), Positives = 59/92 (64%), Gaps = 0/92 (0%)
Query 2 DVTENFFYRPTTRVTNQVYEQLLVVLQQQLGDRPDEVLRGAADEVLQVLKEDGLKEGERK 61
D+++ Y P++ T YE++L +LQ+ L D+P +VLR A EV+ L+ GLK +R+
Sbjct 95 DISDVNHYTPSSVSTRVKYEEILGILQECLTDQPHDVLRDAFGEVMTHLRSPGLKSEDRR 154
Query 62 RGVEGVLGPITQERFTKLFQLSKSITDFMTAD 93
+ E +LGPIT ++F +L+ SK + DF +D
Sbjct 155 KLCEEILGPITDDQFYRLYHASKDLVDFGDSD 186
> ath:AT1G20960 emb1507 (embryo defective 1507); ATP binding /
ATP-dependent helicase/ helicase/ nucleic acid binding / nucleoside-triphosphatase/
nucleotide binding; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2171
Score = 73.6 bits (179), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 35/86 (40%), Positives = 53/86 (61%), Gaps = 0/86 (0%)
Query 4 TENFFYRPTTRVTNQVYEQLLVVLQQQLGDRPDEVLRGAADEVLQVLKEDGLKEGERKRG 63
T++ Y+P T+ T YE +L ++Q+QLG +P ++ GAADE+L VLK D + E+K
Sbjct 105 TDDAVYQPKTKETRAAYEAMLGLIQKQLGGQPPSIVSGAADEILAVLKNDAFRNPEKKME 164
Query 64 VEGVLGPITQERFTKLFQLSKSITDF 89
+E +L I F +L + K ITDF
Sbjct 165 IEKLLNKIENHEFDQLVSIGKLITDF 190
> hsa:23020 SNRNP200, ASCC3L1, BRR2, FLJ11521, HELIC2, RP33, U5-200KD;
small nuclear ribonucleoprotein 200kDa (U5) (EC:3.6.4.13);
K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2136
Score = 73.6 bits (179), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 36/83 (43%), Positives = 53/83 (63%), Gaps = 0/83 (0%)
Query 7 FFYRPTTRVTNQVYEQLLVVLQQQLGDRPDEVLRGAADEVLQVLKEDGLKEGERKRGVEG 66
Y+P T+ T + YE LL +Q LGD+P ++L GAADEVL VLK + L++ ER++ ++
Sbjct 100 IIYKPKTKETRETYEVLLSFIQAALGDQPRDILCGAADEVLAVLKNEKLRDKERRKEIDL 159
Query 67 VLGPITQERFTKLFQLSKSITDF 89
+LG R+ L L K ITD+
Sbjct 160 LLGQTDDTRYHVLVNLGKKITDY 182
> ath:AT2G42270 U5 small nuclear ribonucleoprotein helicase, putative
Length=2172
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 35/86 (40%), Positives = 57/86 (66%), Gaps = 0/86 (0%)
Query 4 TENFFYRPTTRVTNQVYEQLLVVLQQQLGDRPDEVLRGAADEVLQVLKEDGLKEGERKRG 63
TE+ Y+P T+ T +E +L ++QQQLG +P +++ GAADE+L VLK + +K E+K
Sbjct 105 TEDGVYQPKTKETRVAFEIMLGLIQQQLGGQPLDIVCGAADEILAVLKNESVKNHEKKVE 164
Query 64 VEGVLGPITQERFTKLFQLSKSITDF 89
+E +L IT + F++ + K ITD+
Sbjct 165 IEKLLNVITDQVFSQFVSIGKLITDY 190
> tpv:TP03_0747 ATP-dependent RNA helicase; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2249
Score = 70.1 bits (170), Expect = 2e-12, Method: Composition-based stats.
Identities = 39/91 (42%), Positives = 53/91 (58%), Gaps = 1/91 (1%)
Query 2 DVTENFFYRPTTRVTNQVYEQLLVVLQQQLGDRPDEVLRGAADEVLQVLKEDGLKEGERK 61
D+ Y P + T Y+++L +LQ+ LGD+P VL A DE L LK + E ER+
Sbjct 94 DINVGLNYVPRSGYTRTKYQEMLTILQKILGDQPQSVLLSACDETLLSLKTENSAE-ERR 152
Query 62 RGVEGVLGPITQERFTKLFQLSKSITDFMTA 92
VE VLGP++ E + LF LSK +TDF A
Sbjct 153 ALVEDVLGPVSDEVYYNLFHLSKELTDFKMA 183
> pfa:PFD1060w U5 small nuclear ribonucleoprotein-specific protein,
putative; K01509 adenosinetriphosphatase [EC:3.6.1.3]
Length=2874
Score = 58.5 bits (140), Expect = 5e-09, Method: Composition-based stats.
Identities = 30/89 (33%), Positives = 53/89 (59%), Gaps = 1/89 (1%)
Query 2 DVTENFFYRPTTRVTNQVYEQLLVVLQQQLGDRPDEVLRGAADEVLQVLKEDGLKEGERK 61
++ + F Y+PTT+ T ++Y +L+ ++ LGD +++ A +E+L +LK + L E+K
Sbjct 106 NIEDIFLYKPTTKYTEEIYTKLMSKIRFLLGDNTGDIINSACNEILYILKNEELNNEEKK 165
Query 62 RGVEGVLGP-ITQERFTKLFQLSKSITDF 89
+ VE L I + F ++ LSK I DF
Sbjct 166 KQVESELEIYINDDIFIEINNLSKEIYDF 194
> cpv:cgd5_2960 U5snrp Brr2 SFII RNA helicase (sec63 and the second
part of the RNA
Length=1204
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 26/81 (32%), Positives = 45/81 (55%), Gaps = 0/81 (0%)
Query 9 YRPTTRVTNQVYEQLLVVLQQQLGDRPDEVLRGAADEVLQVLKEDGLKEGERKRGVEGVL 68
Y PTT T Y++LL +L LG +P ++L A E+L VLK D L++ E+ + + ++
Sbjct 94 YTPTTMETRLAYDELLGILTDILGSQPSKILEDTAFELLIVLKNDELRDEEKYKKCKEII 153
Query 69 GPITQERFTKLFQLSKSITDF 89
+ ++ + S+ I DF
Sbjct 154 IQLNNTTYSDILDKSRRIVDF 174
> mmu:545030 Wdfy4; WD repeat and FYVE domain containing 4
Length=3024
Score = 32.3 bits (72), Expect = 0.38, Method: Composition-based stats.
Identities = 18/50 (36%), Positives = 27/50 (54%), Gaps = 1/50 (2%)
Query 9 YRPTTRVTNQVYEQLLVVLQQQLGDRPDEVLRGAADEVLQVLKEDGLKEG 58
+R R+ V +QL +QQ L +P E R AA ++LQ K D ++G
Sbjct 89 FRSLQRLAEDVSDQLAQEIQQALAGKPAEQARAAAGQLLQ-WKSDADQDG 137
> cpv:cgd6_3450 EBNA1 binding protein 2; nucleolar protein p40
; K14823 rRNA-processing protein EBP2
Length=246
Score = 31.6 bits (70), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 23/85 (27%), Positives = 45/85 (52%), Gaps = 10/85 (11%)
Query 5 ENFFYRPTTRVTN---QVYEQLLVVLQQQLGDRPDEVLRG--AADEVLQVLKEDGLKEGE 59
E++FY T R N ++ E++ + +RPD++L D+ ++ +K D LK+ +
Sbjct 81 ESYFYSNTLRNVNKGCEILEKMNINW-----NRPDDMLAEMLKTDQHMKKIKMDLLKQEQ 135
Query 60 RKRGVEGVLGPITQERFTKLFQLSK 84
+ + VE +++FTK Q+ K
Sbjct 136 KIKAVELKKQKYVEKKFTKKIQVEK 160
> dre:100151092 efcab6; EF-hand calcium binding domain 6
Length=1032
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 18/55 (32%), Positives = 30/55 (54%), Gaps = 2/55 (3%)
Query 26 VLQQQLGDRPDEVLRGAADEVLQVLKEDGLKEGERKRGVEGVLGPITQERFTKLF 80
+L +++ DR D++ AA E + +GE +R +EG L P+TQ +F L
Sbjct 80 LLLEKVKDRKDDL--KAAFEAFDQEGNKTVTKGEFRRVIEGFLVPLTQSQFESLL 132
> xla:398697 mtus1, fbx27; microtubule associated tumor suppressor
1
Length=1338
Score = 28.1 bits (61), Expect = 6.9, Method: Composition-based stats.
Identities = 23/90 (25%), Positives = 42/90 (46%), Gaps = 12/90 (13%)
Query 16 TNQVYEQLLVVLQQQLGDRPDEV-LRGAADEVLQVLKEDGL-----------KEGERKRG 63
+NQ +E L VV+QQ + R + + R A + L L+ D + ++ E +
Sbjct 991 SNQRFEALTVVVQQLINQREETLKKRKALSQELLNLRGDLVCASSTCERLEKEKNELLKA 1050
Query 64 VEGVLGPITQERFTKLFQLSKSITDFMTAD 93
EG+L + +E +L L + + F T +
Sbjct 1051 YEGILQKVKEEHHAELSDLEEKLKQFYTGE 1080
Lambda K H
0.317 0.137 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2074765676
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40