bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2854_orf1
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
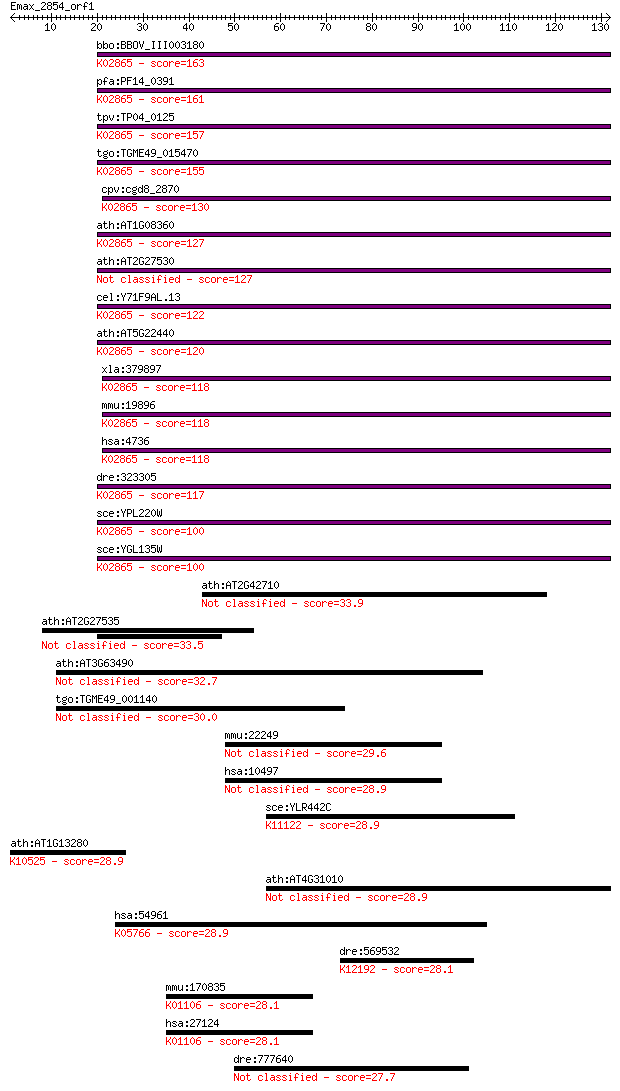
bbo:BBOV_III003180 17.m07305; ribosomal protein L1; K02865 lar... 163 1e-40
pfa:PF14_0391 60S ribosomal protein L1, putative; K02865 large... 161 5e-40
tpv:TP04_0125 60S ribosomal protein L1; K02865 large subunit r... 157 7e-39
tgo:TGME49_015470 60S ribosomal protein L10a, putative ; K0286... 155 4e-38
cpv:cgd8_2870 60S ribosomal protein L10A ; K02865 large subuni... 130 1e-30
ath:AT1G08360 60S ribosomal protein L10A (RPL10aA); K02865 lar... 127 1e-29
ath:AT2G27530 PGY1; PGY1 (PIGGYBACK1); RNA binding / structura... 127 1e-29
cel:Y71F9AL.13 rpl-1; Ribosomal Protein, Large subunit family ... 122 4e-28
ath:AT5G22440 60S ribosomal protein L10A (RPL10aC); K02865 lar... 120 8e-28
xla:379897 rpl10a, MGC53614; ribosomal protein L10a (EC:3.6.5.... 118 4e-27
mmu:19896 Rpl10a, CsA-19, Nedd6; ribosomal protein L10A (EC:3.... 118 6e-27
hsa:4736 RPL10A, Csa-19, NEDD6; ribosomal protein L10a; K02865... 118 6e-27
dre:323305 rpl10a, MGC86881, wu:fb94f08, zgc:73082, zgc:86881;... 117 1e-26
sce:YPL220W RPL1A, SSM1; L1A; K02865 large subunit ribosomal p... 100 2e-21
sce:YGL135W RPL1B, SSM2; L1B; K02865 large subunit ribosomal p... 100 2e-21
ath:AT2G42710 ribosomal protein L1 family protein 33.9 0.15
ath:AT2G27535 ribosomal protein L10A family protein 33.5 0.19
ath:AT3G63490 ribosomal protein L1 family protein 32.7 0.34
tgo:TGME49_001140 proline rich protein, putative 30.0 1.9
mmu:22249 Unc13b, Munc13-1, Munc13-2, Unc13a, Unc13h1, Unc13h2... 29.6 2.5
hsa:10497 UNC13B, MGC133279, MGC133280, MUNC13, UNC13, Unc13h2... 28.9 3.8
sce:YLR442C SIR3, CMT1, MAR2, STE8; Silencing protein that int... 28.9 3.9
ath:AT1G13280 AOC4; AOC4 (ALLENE OXIDE CYCLASE 4); allene-oxid... 28.9 4.0
ath:AT4G31010 RNA binding 28.9 4.2
hsa:54961 SSH3, FLJ10928, FLJ20515, FLJ42240, SSH3L; slingshot... 28.9 4.3
dre:569532 novel protein similar to H.sapiens CHMP2B, chromati... 28.1 7.1
mmu:170835 Inpp5j, Pib5pa, Pipp; inositol polyphosphate 5-phos... 28.1 7.8
hsa:27124 INPP5J, INPP5, MGC129984, PIB5PA, PIPP; inositol pol... 28.1 7.8
dre:777640 bicdr2, ccdc64b; zgc:153929 27.7 8.6
> bbo:BBOV_III003180 17.m07305; ribosomal protein L1; K02865 large
subunit ribosomal protein L10Ae
Length=217
Score = 163 bits (413), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 78/112 (69%), Positives = 94/112 (83%), Gaps = 0/112 (0%)
Query 20 MSKLSQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRA 79
MSKL+ ETLN AI++IL SQEKKR FVET+ELQISLKDYDTQRDKRFSG+V L +VPR
Sbjct 1 MSKLTVETLNEAISAILTQSQEKKRNFVETVELQISLKDYDTQRDKRFSGTVVLQNVPRE 60
Query 80 KSKVCVIGDAVHMEEAKALGLDCMDIEALKKINKNKKIVKKLARKYDAFLAS 131
+ K+CV GD VH ++AK+LG+D +D+E LKK N+NK +VKKLA KY AFLAS
Sbjct 61 RMKICVFGDQVHCDQAKSLGIDYIDLEGLKKFNRNKTLVKKLANKYGAFLAS 112
> pfa:PF14_0391 60S ribosomal protein L1, putative; K02865 large
subunit ribosomal protein L10Ae
Length=217
Score = 161 bits (407), Expect = 5e-40, Method: Compositional matrix adjust.
Identities = 78/112 (69%), Positives = 97/112 (86%), Gaps = 0/112 (0%)
Query 20 MSKLSQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRA 79
MSKL+Q+ L AI+ + +G+++KKRKFVETIELQI LKDYDTQRDKRFSG+VRL + R
Sbjct 1 MSKLNQDLLKKAISDVFEGTKQKKRKFVETIELQIGLKDYDTQRDKRFSGTVRLSNEVRK 60
Query 80 KSKVCVIGDAVHMEEAKALGLDCMDIEALKKINKNKKIVKKLARKYDAFLAS 131
K KVC++GDAVH+EEA+ L LD MDIEA+KK+NK+K +VKKLA+KYDAFLAS
Sbjct 61 KLKVCILGDAVHVEEAQKLELDYMDIEAMKKLNKDKTLVKKLAKKYDAFLAS 112
> tpv:TP04_0125 60S ribosomal protein L1; K02865 large subunit
ribosomal protein L10Ae
Length=217
Score = 157 bits (397), Expect = 7e-39, Method: Compositional matrix adjust.
Identities = 75/112 (66%), Positives = 91/112 (81%), Gaps = 0/112 (0%)
Query 20 MSKLSQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRA 79
MSKLS + LN A+ ++L+GS+ KKR F ET+ELQISLKDYDTQRDKRFSG+V L + P+
Sbjct 1 MSKLSSDRLNDAVTAVLEGSKTKKRNFTETVELQISLKDYDTQRDKRFSGTVVLQNAPKK 60
Query 80 KSKVCVIGDAVHMEEAKALGLDCMDIEALKKINKNKKIVKKLARKYDAFLAS 131
KVCV GDAVH +EAKALG+D +D+E LKK N+NK +VKKLA KY AFLAS
Sbjct 61 NMKVCVFGDAVHCDEAKALGVDFIDLEGLKKFNRNKTLVKKLANKYSAFLAS 112
> tgo:TGME49_015470 60S ribosomal protein L10a, putative ; K02865
large subunit ribosomal protein L10Ae
Length=217
Score = 155 bits (391), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 86/112 (76%), Positives = 100/112 (89%), Gaps = 0/112 (0%)
Query 20 MSKLSQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRA 79
MSKLS + L AI IL+GS+EKKRKFVET+ELQI LKDYDTQRDKRFSGSVRLP+VPR
Sbjct 1 MSKLSTDGLKKAIGEILEGSREKKRKFVETVELQIGLKDYDTQRDKRFSGSVRLPNVPRP 60
Query 80 KSKVCVIGDAVHMEEAKALGLDCMDIEALKKINKNKKIVKKLARKYDAFLAS 131
+ +VCV+GDAVH E+AK LGL+ MD+EA+KK+NKNKK+VKKLARKYDAFLAS
Sbjct 61 RMRVCVMGDAVHCEQAKELGLEFMDVEAMKKLNKNKKLVKKLARKYDAFLAS 112
> cpv:cgd8_2870 60S ribosomal protein L10A ; K02865 large subunit
ribosomal protein L10Ae
Length=221
Score = 130 bits (327), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 75/111 (67%), Positives = 91/111 (81%), Gaps = 0/111 (0%)
Query 21 SKLSQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRAK 80
K+S ET+ AI+ IL+GS+ K RKFVET+ELQI LKDYDTQRDKRF+G+VRLP+VPR
Sbjct 6 GKVSSETIRRAISEILEGSKAKPRKFVETVELQIGLKDYDTQRDKRFAGTVRLPNVPRPN 65
Query 81 SKVCVIGDAVHMEEAKALGLDCMDIEALKKINKNKKIVKKLARKYDAFLAS 131
++VCV+GDA E A+ LG D MDIE +KKINKNKK+VKKL +KYD FLAS
Sbjct 66 ARVCVMGDAADCERAQKLGFDVMDIEEMKKINKNKKVVKKLCKKYDLFLAS 116
> ath:AT1G08360 60S ribosomal protein L10A (RPL10aA); K02865 large
subunit ribosomal protein L10Ae
Length=216
Score = 127 bits (319), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 74/112 (66%), Positives = 93/112 (83%), Gaps = 0/112 (0%)
Query 20 MSKLSQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRA 79
MSKL E + AI +I S+ KKR FVETIELQI LK+YD Q+DKRFSGSV+LPH+PR
Sbjct 1 MSKLQSEAVREAITTITGKSEAKKRNFVETIELQIGLKNYDPQKDKRFSGSVKLPHIPRP 60
Query 80 KSKVCVIGDAVHMEEAKALGLDCMDIEALKKINKNKKIVKKLARKYDAFLAS 131
K K+C++GDA H+EEA+ +GL+ MD+E+LKK+NKNKK+VKKLA+KY AFLAS
Sbjct 61 KMKICMLGDAQHVEEAEKMGLENMDVESLKKLNKNKKLVKKLAKKYHAFLAS 112
> ath:AT2G27530 PGY1; PGY1 (PIGGYBACK1); RNA binding / structural
constituent of ribosome
Length=216
Score = 127 bits (318), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 74/112 (66%), Positives = 92/112 (82%), Gaps = 0/112 (0%)
Query 20 MSKLSQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRA 79
MSKL E + AI +I S+EKKR FVET+ELQI LK+YD Q+DKRFSGSV+LPH+PR
Sbjct 1 MSKLQSEAVREAITTIKGKSEEKKRNFVETVELQIGLKNYDPQKDKRFSGSVKLPHIPRP 60
Query 80 KSKVCVIGDAVHMEEAKALGLDCMDIEALKKINKNKKIVKKLARKYDAFLAS 131
K K+C++GDA H+EEA+ +GL MD+EALKK+NKNKK+VKKLA+ Y AFLAS
Sbjct 61 KMKICMLGDAQHVEEAEKMGLSNMDVEALKKLNKNKKLVKKLAKSYHAFLAS 112
> cel:Y71F9AL.13 rpl-1; Ribosomal Protein, Large subunit family
member (rpl-1); K02865 large subunit ribosomal protein L10Ae
Length=216
Score = 122 bits (305), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 71/112 (63%), Positives = 88/112 (78%), Gaps = 0/112 (0%)
Query 20 MSKLSQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRA 79
MSK+S+E+LN AIA +LKGS EK RKF ETIELQI LK+YD Q+DKRFSGS+RL H+PR
Sbjct 1 MSKVSRESLNEAIAEVLKGSSEKPRKFRETIELQIGLKNYDPQKDKRFSGSIRLKHIPRP 60
Query 80 KSKVCVIGDAVHMEEAKALGLDCMDIEALKKINKNKKIVKKLARKYDAFLAS 131
KVCV GD H++EA A + M + LKK+NK KK++KKLA+ YDAF+AS
Sbjct 61 NMKVCVFGDQHHLDEAAAGDIPSMSADDLKKLNKQKKLIKKLAKSYDAFIAS 112
> ath:AT5G22440 60S ribosomal protein L10A (RPL10aC); K02865 large
subunit ribosomal protein L10Ae
Length=217
Score = 120 bits (302), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 74/113 (65%), Positives = 93/113 (82%), Gaps = 1/113 (0%)
Query 20 MSKLSQETLNAAIASILKGSQE-KKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPR 78
MSKL E + AI+SI+ +E K R F ETIELQI LK+YD Q+DKRFSGSV+LPHVPR
Sbjct 1 MSKLQSEAVREAISSIITHCKETKPRNFTETIELQIGLKNYDPQKDKRFSGSVKLPHVPR 60
Query 79 AKSKVCVIGDAVHMEEAKALGLDCMDIEALKKINKNKKIVKKLARKYDAFLAS 131
K K+C++GDA H+EEA+ +GL+ MD+EALKK+NKNKK+VKKLA+K+ AFLAS
Sbjct 61 PKMKICMLGDAQHVEEAEKIGLESMDVEALKKLNKNKKLVKKLAKKFHAFLAS 113
> xla:379897 rpl10a, MGC53614; ribosomal protein L10a (EC:3.6.5.3);
K02865 large subunit ribosomal protein L10Ae
Length=217
Score = 118 bits (296), Expect = 4e-27, Method: Compositional matrix adjust.
Identities = 74/111 (66%), Positives = 94/111 (84%), Gaps = 0/111 (0%)
Query 21 SKLSQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRAK 80
SK+S++TL A+ +L+GS++KKRKF+ET+ELQISLK+YD Q+DKRFSG+VRL PR K
Sbjct 3 SKVSRDTLYEAVREVLQGSKKKKRKFLETVELQISLKNYDPQKDKRFSGTVRLKSTPRPK 62
Query 81 SKVCVIGDAVHMEEAKALGLDCMDIEALKKINKNKKIVKKLARKYDAFLAS 131
+CV+GD H +EAKA+ L MDIEALKK+NKNKK+VKKLA+KYDAFLAS
Sbjct 63 FSLCVLGDQQHCDEAKAVDLPHMDIEALKKLNKNKKLVKKLAKKYDAFLAS 113
> mmu:19896 Rpl10a, CsA-19, Nedd6; ribosomal protein L10A (EC:3.6.5.3);
K02865 large subunit ribosomal protein L10Ae
Length=217
Score = 118 bits (295), Expect = 6e-27, Method: Compositional matrix adjust.
Identities = 73/111 (65%), Positives = 92/111 (82%), Gaps = 0/111 (0%)
Query 21 SKLSQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRAK 80
SK+S++TL A+ +L G+Q K+RKF+ET+ELQISLK+YD Q+DKRFSG+VRL PR K
Sbjct 3 SKVSRDTLYEAVREVLHGNQRKRRKFLETVELQISLKNYDPQKDKRFSGTVRLKSTPRPK 62
Query 81 SKVCVIGDAVHMEEAKALGLDCMDIEALKKINKNKKIVKKLARKYDAFLAS 131
VCV+GD H +EAKA+ + MDIEALKK+NKNKK+VKKLA+KYDAFLAS
Sbjct 63 FSVCVLGDQQHCDEAKAVDIPHMDIEALKKLNKNKKLVKKLAKKYDAFLAS 113
> hsa:4736 RPL10A, Csa-19, NEDD6; ribosomal protein L10a; K02865
large subunit ribosomal protein L10Ae
Length=217
Score = 118 bits (295), Expect = 6e-27, Method: Compositional matrix adjust.
Identities = 73/111 (65%), Positives = 92/111 (82%), Gaps = 0/111 (0%)
Query 21 SKLSQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRAK 80
SK+S++TL A+ +L G+Q K+RKF+ET+ELQISLK+YD Q+DKRFSG+VRL PR K
Sbjct 3 SKVSRDTLYEAVREVLHGNQRKRRKFLETVELQISLKNYDPQKDKRFSGTVRLKSTPRPK 62
Query 81 SKVCVIGDAVHMEEAKALGLDCMDIEALKKINKNKKIVKKLARKYDAFLAS 131
VCV+GD H +EAKA+ + MDIEALKK+NKNKK+VKKLA+KYDAFLAS
Sbjct 63 FSVCVLGDQQHCDEAKAVDIPHMDIEALKKLNKNKKLVKKLAKKYDAFLAS 113
> dre:323305 rpl10a, MGC86881, wu:fb94f08, zgc:73082, zgc:86881;
ribosomal protein L10a; K02865 large subunit ribosomal protein
L10Ae
Length=216
Score = 117 bits (292), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 75/112 (66%), Positives = 91/112 (81%), Gaps = 0/112 (0%)
Query 20 MSKLSQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRA 79
MSK+S++TL A+ + GS+ KKRKF+ET+ELQISLK+YD Q+DKRFSG+VRL PR
Sbjct 1 MSKVSRDTLYEAVKEVQAGSRRKKRKFLETVELQISLKNYDPQKDKRFSGTVRLKTTPRP 60
Query 80 KSKVCVIGDAVHMEEAKALGLDCMDIEALKKINKNKKIVKKLARKYDAFLAS 131
K VCV+GD H +EAKA L MDIEALKK+NKNKK+VKKLA+KYDAFLAS
Sbjct 61 KFSVCVLGDQQHCDEAKAAELPHMDIEALKKLNKNKKLVKKLAKKYDAFLAS 112
> sce:YPL220W RPL1A, SSM1; L1A; K02865 large subunit ribosomal
protein L10Ae
Length=217
Score = 100 bits (248), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 59/113 (52%), Positives = 86/113 (76%), Gaps = 1/113 (0%)
Query 20 MSKLSQETLNAAIASILKGSQE-KKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPR 78
MSK++ + + +LK S E KKR F+ET+ELQ+ LK+YD QRDKRFSGS++LP+ PR
Sbjct 1 MSKITSSQVREHVKELLKYSNETKKRNFLETVELQVGLKNYDPQRDKRFSGSLKLPNCPR 60
Query 79 AKSKVCVIGDAVHMEEAKALGLDCMDIEALKKINKNKKIVKKLARKYDAFLAS 131
+C+ GDA ++ AK+ G+D M ++ LKK+NKNKK++KKL++KY+AF+AS
Sbjct 61 PNMSICIFGDAFDVDRAKSCGVDAMSVDDLKKLNKNKKLIKKLSKKYNAFIAS 113
> sce:YGL135W RPL1B, SSM2; L1B; K02865 large subunit ribosomal
protein L10Ae
Length=217
Score = 100 bits (248), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 59/113 (52%), Positives = 86/113 (76%), Gaps = 1/113 (0%)
Query 20 MSKLSQETLNAAIASILKGSQE-KKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPR 78
MSK++ + + +LK S E KKR F+ET+ELQ+ LK+YD QRDKRFSGS++LP+ PR
Sbjct 1 MSKITSSQVREHVKELLKYSNETKKRNFLETVELQVGLKNYDPQRDKRFSGSLKLPNCPR 60
Query 79 AKSKVCVIGDAVHMEEAKALGLDCMDIEALKKINKNKKIVKKLARKYDAFLAS 131
+C+ GDA ++ AK+ G+D M ++ LKK+NKNKK++KKL++KY+AF+AS
Sbjct 61 PNMSICIFGDAFDVDRAKSCGVDAMSVDDLKKLNKNKKLIKKLSKKYNAFIAS 113
> ath:AT2G42710 ribosomal protein L1 family protein
Length=415
Score = 33.9 bits (76), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 24/76 (31%), Positives = 39/76 (51%), Gaps = 1/76 (1%)
Query 43 KRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRAKSKVCVIGDAVHMEEAKALGLDC 102
K KF ET+E + L + + G++ LPH + KV + E+AKA G D
Sbjct 202 KAKFDETLEAHVRLGIEKGRSELIVRGTLALPHSVKKDVKVAFFAEGADAEDAKAAGADV 261
Query 103 M-DIEALKKINKNKKI 117
+ +E +++I K+ KI
Sbjct 262 VGGLELIEEILKSGKI 277
> ath:AT2G27535 ribosomal protein L10A family protein
Length=81
Score = 33.5 bits (75), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 28/46 (60%), Gaps = 0/46 (0%)
Query 8 TLCRTAASKYTKMSKLSQETLNAAIASILKGSQEKKRKFVETIELQ 53
+ R + + KM +L E+L AIA+I S+EKKR +T++LQ
Sbjct 15 NITRPSEERKLKMRQLESESLREAIATIEGKSEEKKRNVGKTVKLQ 60
Score = 28.1 bits (61), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 14/27 (51%), Positives = 20/27 (74%), Gaps = 0/27 (0%)
Query 20 MSKLSQETLNAAIASILKGSQEKKRKF 46
MSKL+ +TL AIA+I + S+E+K K
Sbjct 1 MSKLNSDTLREAIANITRPSEERKLKM 27
> ath:AT3G63490 ribosomal protein L1 family protein
Length=346
Score = 32.7 bits (73), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 27/97 (27%), Positives = 46/97 (47%), Gaps = 7/97 (7%)
Query 11 RTAASKYTKMSKLSQE----TLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKR 66
RT + ++ ++ KL + +N AI S+LK Q +FVE++E L D++
Sbjct 110 RTRSKRFLEIQKLRETKKEYDVNTAI-SLLK--QTANTRFVESVEAHFRLNIDPKYNDQQ 166
Query 67 FSGSVRLPHVPRAKSKVCVIGDAVHMEEAKALGLDCM 103
+V LP V V+ ++EAK+ G D +
Sbjct 167 LRATVSLPKGTGQTVIVAVLAQGEKVDEAKSAGADIV 203
> tgo:TGME49_001140 proline rich protein, putative
Length=1051
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 22/68 (32%), Positives = 35/68 (51%), Gaps = 6/68 (8%)
Query 11 RTAASKYTKMSKLSQETLNAAIASIL-----KGSQEKKRKFVETIELQISLKDYDTQRDK 65
+ A K ++KL+ E+L AIA + + +E R + +I+ +ISL D Q+D
Sbjct 753 QCAKKKQELLAKLT-ESLKEAIARLTDAQTPENEKEAVRAMIASIKEKISLLSDDKQKDA 811
Query 66 RFSGSVRL 73
R VRL
Sbjct 812 RPEEEVRL 819
> mmu:22249 Unc13b, Munc13-1, Munc13-2, Unc13a, Unc13h1, Unc13h2;
unc-13 homolog B (C. elegans)
Length=1602
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 24/51 (47%), Gaps = 4/51 (7%)
Query 48 ETIELQISLKDYDTQRDKRFSGSVRLPHVPRAKSKVCV----IGDAVHMEE 94
E ELQI +KDY R+ R G +P A C +G +HM+E
Sbjct 1517 EAYELQICVKDYCFAREDRVIGLAVMPLRDVAAKGSCACWCPLGRKIHMDE 1567
> hsa:10497 UNC13B, MGC133279, MGC133280, MUNC13, UNC13, Unc13h2,
hmunc13; unc-13 homolog B (C. elegans)
Length=1591
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 19/51 (37%), Positives = 26/51 (50%), Gaps = 4/51 (7%)
Query 48 ETIELQISLKDYDTQRDKRFSGSVRLP-HVPRAKSKV---CVIGDAVHMEE 94
E+ ELQI +KDY R+ R G +P AK C +G +HM+E
Sbjct 1506 ESYELQICVKDYCFAREDRVLGLAVMPLRDVTAKGSCACWCPLGRKIHMDE 1556
> sce:YLR442C SIR3, CMT1, MAR2, STE8; Silencing protein that interacts
with Sir2p and Sir4p, and histone H3 and H4 tails,
to establish a transcriptionally silent chromatin state; required
for spreading of silenced chromatin; recruited to chromatin
through interaction with Rap1p; K11122 regulatory protein
SIR3
Length=978
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 15/54 (27%), Positives = 28/54 (51%), Gaps = 1/54 (1%)
Query 57 KDYDTQRDKRFSGSVRLPHVPRAKSKVCVIGDAVHMEEAKALGLDCMDIEALKK 110
K+Y T+ D +G+ + P +PR +K+ + + E+ A +D + LKK
Sbjct 421 KNYTTETDNEMNGNGK-PGIPRGNTKIHSMNENPTPEKGNAKMIDFATLSKLKK 473
> ath:AT1G13280 AOC4; AOC4 (ALLENE OXIDE CYCLASE 4); allene-oxide
cyclase (EC:5.3.99.6); K10525 allene oxide cyclase [EC:5.3.99.6]
Length=254
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 8/25 (32%), Positives = 20/25 (80%), Gaps = 0/25 (0%)
Query 1 ASGGIFRTLCRTAASKYTKMSKLSQ 25
++GG FRT+C ++++ Y++ +K+ +
Sbjct 59 STGGFFRTICSSSSNDYSRPTKIQE 83
> ath:AT4G31010 RNA binding
Length=405
Score = 28.9 bits (63), Expect = 4.2, Method: Composition-based stats.
Identities = 19/75 (25%), Positives = 34/75 (45%), Gaps = 0/75 (0%)
Query 57 KDYDTQRDKRFSGSVRLPHVPRAKSKVCVIGDAVHMEEAKALGLDCMDIEALKKINKNKK 116
++YD ++ + + PH P + D + ++E KA+ + + AL K+ KN
Sbjct 246 RNYDPKKRPKIPLMLWKPHEPVYPRLIKTTIDGLSIDETKAMRKKGLAVPALTKLAKNGY 305
Query 117 IVKKLARKYDAFLAS 131
+ DAFL S
Sbjct 306 YGSLVPMVRDAFLVS 320
> hsa:54961 SSH3, FLJ10928, FLJ20515, FLJ42240, SSH3L; slingshot
homolog 3 (Drosophila) (EC:3.1.3.16 3.1.3.48); K05766 slingshot
Length=659
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 20/84 (23%), Positives = 38/84 (45%), Gaps = 3/84 (3%)
Query 24 SQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRAKSKV 83
S+E L+ +GSQ +++ + L + ++ Q D R + + P PR + +
Sbjct 70 SEEELHGDQTDFGQGSQSPQKQEEQRQHLHLMVQLLRPQDDIRLAAQLEAPRPPRLRYLL 129
Query 84 CVI---GDAVHMEEAKALGLDCMD 104
V G+ + +E LG+D D
Sbjct 130 VVSTREGEGLSQDETVLLGVDFPD 153
> dre:569532 novel protein similar to H.sapiens CHMP2B, chromatin
modifying protein 2B (CHMP2B); K12192 charged multivesicular
body protein 2B
Length=216
Score = 28.1 bits (61), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 14/29 (48%), Positives = 16/29 (55%), Gaps = 0/29 (0%)
Query 73 LPHVPRAKSKVCVIGDAVHMEEAKALGLD 101
LP AKSK I DA + KALG+D
Sbjct 188 LPGASPAKSKTATISDAEIERQLKALGMD 216
> mmu:170835 Inpp5j, Pib5pa, Pipp; inositol polyphosphate 5-phosphatase
J (EC:3.1.3.56); K01106 inositol-1,4,5-trisphosphate
5-phosphatase [EC:3.1.3.56]
Length=1003
Score = 28.1 bits (61), Expect = 7.8, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 15/32 (46%), Gaps = 0/32 (0%)
Query 35 ILKGSQEKKRKFVETIELQISLKDYDTQRDKR 66
ILKG QE F T + + YDT KR
Sbjct 640 ILKGFQEGPLNFAPTFKFDVGTNKYDTSAKKR 671
> hsa:27124 INPP5J, INPP5, MGC129984, PIB5PA, PIPP; inositol polyphosphate-5-phosphatase
J (EC:3.1.3.56); K01106 inositol-1,4,5-trisphosphate
5-phosphatase [EC:3.1.3.56]
Length=638
Score = 28.1 bits (61), Expect = 7.8, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 15/32 (46%), Gaps = 0/32 (0%)
Query 35 ILKGSQEKKRKFVETIELQISLKDYDTQRDKR 66
ILKG QE F T + + YDT KR
Sbjct 275 ILKGFQEGPLNFAPTFKFDVGTNKYDTSAKKR 306
> dre:777640 bicdr2, ccdc64b; zgc:153929
Length=446
Score = 27.7 bits (60), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 28/52 (53%), Gaps = 1/52 (1%)
Query 50 IELQISLKDYDTQRDKRFSGSVRLPHVPRA-KSKVCVIGDAVHMEEAKALGL 100
IELQ SLKD +T+ ++ S +L V R + +V +G+ + EA L
Sbjct 236 IELQTSLKDKETELEQEHSTVFQLRTVNRTLQQRVQALGEEASLGEATCFPL 287
Lambda K H
0.317 0.130 0.351
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2105161088
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40