bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2847_orf1
Length=80
Score E
Sequences producing significant alignments: (Bits) Value
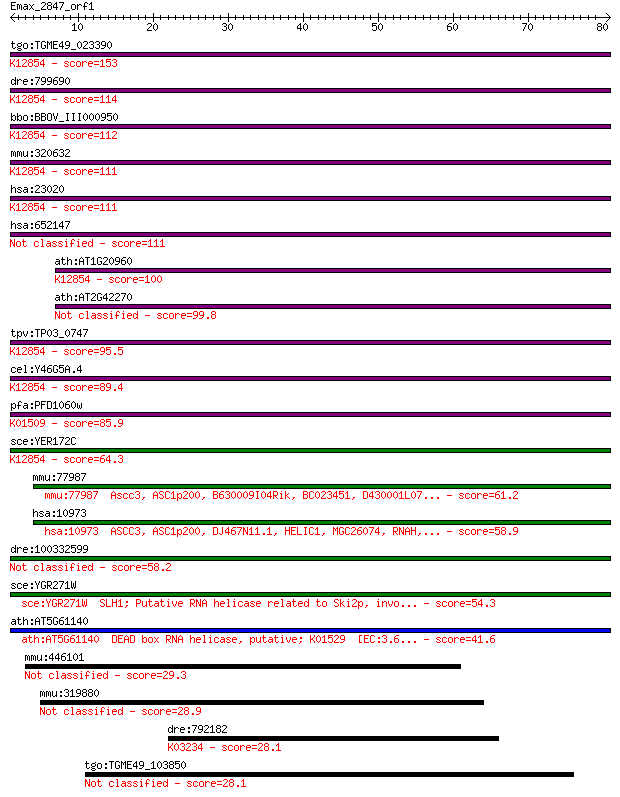
tgo:TGME49_023390 sec63 domain-containing DEAD/DEAH box helica... 153 1e-37
dre:799690 fb63a09; wu:fb63a09 (EC:3.6.4.13); K12854 pre-mRNA-... 114 1e-25
bbo:BBOV_III000950 17.m07111; sec63 domain containing protein ... 112 3e-25
mmu:320632 Snrnp200, A330064G03Rik, Ascc3l1, BC011390, HELIC2,... 111 6e-25
hsa:23020 SNRNP200, ASCC3L1, BRR2, FLJ11521, HELIC2, RP33, U5-... 111 6e-25
hsa:652147 u5 small nuclear ribonucleoprotein 200 kDa helicase... 111 7e-25
ath:AT1G20960 emb1507 (embryo defective 1507); ATP binding / A... 100 9e-22
ath:AT2G42270 U5 small nuclear ribonucleoprotein helicase, put... 99.8 2e-21
tpv:TP03_0747 ATP-dependent RNA helicase; K12854 pre-mRNA-spli... 95.5 4e-20
cel:Y46G5A.4 hypothetical protein; K12854 pre-mRNA-splicing he... 89.4 2e-18
pfa:PFD1060w U5 small nuclear ribonucleoprotein-specific prote... 85.9 3e-17
sce:YER172C BRR2, PRP44, RSS1, SLT22, SNU246; Brr2p (EC:3.6.1.... 64.3 8e-11
mmu:77987 Ascc3, ASC1p200, B630009I04Rik, BC023451, D430001L07... 61.2 8e-10
hsa:10973 ASCC3, ASC1p200, DJ467N11.1, HELIC1, MGC26074, RNAH,... 58.9 4e-09
dre:100332599 mutagen-sensitive 308-like 58.2 7e-09
sce:YGR271W SLH1; Putative RNA helicase related to Ski2p, invo... 54.3 9e-08
ath:AT5G61140 DEAD box RNA helicase, putative; K01529 [EC:3.6... 41.6 6e-04
mmu:446101 Xrra1, AI449753; X-ray radiation resistance associa... 29.3 3.3
mmu:319880 Tmcc3, AW488095, C630016B22Rik, C88213, MGC30586, T... 28.9 3.8
dre:792182 eef2a.1, MGC113191, si:dkey-110c1.3, zgc:113191; eu... 28.1 6.8
tgo:TGME49_103850 hypothetical protein 28.1 7.4
> tgo:TGME49_023390 sec63 domain-containing DEAD/DEAH box helicase,
putative ; K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2198
Score = 153 bits (386), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 72/80 (90%), Positives = 76/80 (95%), Gaps = 0/80 (0%)
Query 1 CNEIQGRMWSTMTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLL 60
C EI RMWS+MTPLRQFKVLPEELLRKIEKKDLPF+RYYDL+STEIGELVRVPKMGKLL
Sbjct 1169 CKEIDRRMWSSMTPLRQFKVLPEELLRKIEKKDLPFERYYDLSSTEIGELVRVPKMGKLL 1228
Query 61 HRLIHSFPKLELAAFVQPLS 80
HRLIH FPKLELAAFVQPL+
Sbjct 1229 HRLIHQFPKLELAAFVQPLT 1248
> dre:799690 fb63a09; wu:fb63a09 (EC:3.6.4.13); K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2134
Score = 114 bits (284), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 47/80 (58%), Positives = 64/80 (80%), Gaps = 0/80 (0%)
Query 1 CNEIQGRMWSTMTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLL 60
C I RMW +M+PLRQF+ LPEE+++KIEKK+ PF+R YDL EIGEL+R+PKMGK +
Sbjct 1112 CKMIDKRMWQSMSPLRQFRKLPEEVIKKIEKKNFPFERLYDLNHNEIGELIRMPKMGKTI 1171
Query 61 HRLIHSFPKLELAAFVQPLS 80
H+ +H FPKL+LA +QP++
Sbjct 1172 HKYVHQFPKLDLAVHLQPIT 1191
> bbo:BBOV_III000950 17.m07111; sec63 domain containing protein
(EC:3.6.1.-); K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2133
Score = 112 bits (280), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 50/80 (62%), Positives = 62/80 (77%), Gaps = 0/80 (0%)
Query 1 CNEIQGRMWSTMTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLL 60
C ++ RMWS M PLRQFK LPEEL+ K+E+ D +DRYYDL+S E+GEL R PK+GK L
Sbjct 1117 CKMVERRMWSVMLPLRQFKSLPEELILKLERNDFTWDRYYDLSSVELGELCRQPKLGKTL 1176
Query 61 HRLIHSFPKLELAAFVQPLS 80
HRL+H P+LEL FVQPL+
Sbjct 1177 HRLVHLVPRLELQVFVQPLT 1196
> mmu:320632 Snrnp200, A330064G03Rik, Ascc3l1, BC011390, HELIC2,
KIAA0788, U5-200-KD, U5-200KD; small nuclear ribonucleoprotein
200 (U5) (EC:3.6.4.13); K12854 pre-mRNA-splicing helicase
BRR2 [EC:3.6.4.13]
Length=2136
Score = 111 bits (277), Expect = 6e-25, Method: Compositional matrix adjust.
Identities = 47/80 (58%), Positives = 63/80 (78%), Gaps = 0/80 (0%)
Query 1 CNEIQGRMWSTMTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLL 60
C I RMW +M PLRQF+ LPEE+++KIEKK+ PF+R YDL EIGEL+R+PKMGK +
Sbjct 1115 CKMIDKRMWQSMCPLRQFRKLPEEVVKKIEKKNFPFERLYDLNHNEIGELIRMPKMGKTI 1174
Query 61 HRLIHSFPKLELAAFVQPLS 80
H+ +H FPKLEL+ +QP++
Sbjct 1175 HKYVHLFPKLELSVHLQPIT 1194
> hsa:23020 SNRNP200, ASCC3L1, BRR2, FLJ11521, HELIC2, RP33, U5-200KD;
small nuclear ribonucleoprotein 200kDa (U5) (EC:3.6.4.13);
K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2136
Score = 111 bits (277), Expect = 6e-25, Method: Compositional matrix adjust.
Identities = 47/80 (58%), Positives = 63/80 (78%), Gaps = 0/80 (0%)
Query 1 CNEIQGRMWSTMTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLL 60
C I RMW +M PLRQF+ LPEE+++KIEKK+ PF+R YDL EIGEL+R+PKMGK +
Sbjct 1115 CKMIDKRMWQSMCPLRQFRKLPEEVVKKIEKKNFPFERLYDLNHNEIGELIRMPKMGKTI 1174
Query 61 HRLIHSFPKLELAAFVQPLS 80
H+ +H FPKLEL+ +QP++
Sbjct 1175 HKYVHLFPKLELSVHLQPIT 1194
> hsa:652147 u5 small nuclear ribonucleoprotein 200 kDa helicase-like
Length=1700
Score = 111 bits (277), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 47/80 (58%), Positives = 63/80 (78%), Gaps = 0/80 (0%)
Query 1 CNEIQGRMWSTMTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLL 60
C I RMW +M PLRQF+ LPEE+++KIEKK+ PF+R YDL EIGEL+R+PKMGK +
Sbjct 679 CKMIDKRMWQSMCPLRQFRKLPEEVVKKIEKKNFPFERLYDLNHNEIGELIRMPKMGKTI 738
Query 61 HRLIHSFPKLELAAFVQPLS 80
H+ +H FPKLEL+ +QP++
Sbjct 739 HKYVHLFPKLELSVHLQPIT 758
> ath:AT1G20960 emb1507 (embryo defective 1507); ATP binding /
ATP-dependent helicase/ helicase/ nucleic acid binding / nucleoside-triphosphatase/
nucleotide binding; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2171
Score = 100 bits (250), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 44/74 (59%), Positives = 59/74 (79%), Gaps = 0/74 (0%)
Query 7 RMWSTMTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHS 66
RMWS TPLRQF L ++L ++EKKDL ++RYYDL++ E+GEL+R PKMGK LH+ IH
Sbjct 1145 RMWSVQTPLRQFHGLSNDILMQLEKKDLVWERYYDLSAQELGELIRSPKMGKPLHKFIHQ 1204
Query 67 FPKLELAAFVQPLS 80
FPK+ L+A VQP++
Sbjct 1205 FPKVTLSAHVQPIT 1218
> ath:AT2G42270 U5 small nuclear ribonucleoprotein helicase, putative
Length=2172
Score = 99.8 bits (247), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 46/74 (62%), Positives = 59/74 (79%), Gaps = 0/74 (0%)
Query 7 RMWSTMTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHS 66
RMWS TPL QF +P+E+L K+EK DL ++RYYDL+S E+GEL+ PKMG+ LH+ IH
Sbjct 1146 RMWSVQTPLWQFPGIPKEILMKLEKNDLVWERYYDLSSQELGELICNPKMGRPLHKYIHQ 1205
Query 67 FPKLELAAFVQPLS 80
FPKL+LAA VQP+S
Sbjct 1206 FPKLKLAAHVQPIS 1219
> tpv:TP03_0747 ATP-dependent RNA helicase; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2249
Score = 95.5 bits (236), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 40/80 (50%), Positives = 57/80 (71%), Gaps = 0/80 (0%)
Query 1 CNEIQGRMWSTMTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLL 60
C ++ RMW M PLR FK +P E++ K+EKKD+P+ RYYDL + E+GEL R K+GK L
Sbjct 1170 CKMVESRMWLLMLPLRHFKTIPNEVVTKLEKKDIPWVRYYDLNAVELGELCRNQKLGKSL 1229
Query 61 HRLIHSFPKLELAAFVQPLS 80
++ +H PK+ L +VQPL+
Sbjct 1230 YKFVHLVPKVNLQVYVQPLT 1249
> cel:Y46G5A.4 hypothetical protein; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2145
Score = 89.4 bits (220), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 36/80 (45%), Positives = 55/80 (68%), Gaps = 0/80 (0%)
Query 1 CNEIQGRMWSTMTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLL 60
C + R W ++ PL QFK +P E++R I+KK+ FDR YDL ++G+L+++PKMGK L
Sbjct 1108 CKMVTQRQWGSLNPLHQFKKIPSEVVRSIDKKNYSFDRLYDLDQHQLGDLIKMPKMGKPL 1167
Query 61 HRLIHSFPKLELAAFVQPLS 80
+ I FPKLE+ +QP++
Sbjct 1168 FKFIRQFPKLEMTTLIQPIT 1187
> pfa:PFD1060w U5 small nuclear ribonucleoprotein-specific protein,
putative; K01509 adenosinetriphosphatase [EC:3.6.1.3]
Length=2874
Score = 85.9 bits (211), Expect = 3e-17, Method: Composition-based stats.
Identities = 38/80 (47%), Positives = 55/80 (68%), Gaps = 0/80 (0%)
Query 1 CNEIQGRMWSTMTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLL 60
C I+ RMWSTMTPLRQF +L EL+R IEKK++ F Y + E + + K+ K +
Sbjct 1513 CKMIERRMWSTMTPLRQFGLLSTELIRIIEKKNITFKNYLTMNLNEYITIFKNKKIAKNI 1572
Query 61 HRLIHSFPKLELAAFVQPLS 80
++L+H FPKLEL A++QP++
Sbjct 1573 YKLVHHFPKLELNAYIQPIN 1592
> sce:YER172C BRR2, PRP44, RSS1, SLT22, SNU246; Brr2p (EC:3.6.1.-);
K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2163
Score = 64.3 bits (155), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 46/81 (56%), Gaps = 1/81 (1%)
Query 1 CNEIQGRMWSTMTPLRQFKVLPEELLRKIEKKDLPFDRYYDL-TSTEIGELVRVPKMGKL 59
C +MW T PLRQFK P E+++++E +P+ Y L T E+G +R K GK
Sbjct 1132 CKSATTKMWPTNCPLRQFKTCPVEVIKRLEASTVPWGDYLQLETPAEVGRAIRSEKYGKQ 1191
Query 60 LHRLIHSFPKLELAAFVQPLS 80
++ L+ FPK+ + QP++
Sbjct 1192 VYDLLKRFPKMSVTCNAQPIT 1212
> mmu:77987 Ascc3, ASC1p200, B630009I04Rik, BC023451, D430001L07Rik,
D630041L21, Helic1, RNAH; activating signal cointegrator
1 complex subunit 3; K01529 [EC:3.6.1.-]
Length=2198
Score = 61.2 bits (147), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 26/77 (33%), Positives = 46/77 (59%), Gaps = 0/77 (0%)
Query 4 IQGRMWSTMTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLLHRL 63
I R+W +PLRQF VLP +L ++E+K+L D+ D+ EIG ++ +G + +
Sbjct 1117 IDKRLWGWASPLRQFSVLPPHILTRLEEKNLTVDKLKDMRKDEIGHILHHVNIGLKVKQC 1176
Query 64 IHSFPKLELAAFVQPLS 80
+H P + + A +QP++
Sbjct 1177 VHQIPSVTMEASIQPIT 1193
> hsa:10973 ASCC3, ASC1p200, DJ467N11.1, HELIC1, MGC26074, RNAH,
dJ121G13.4; activating signal cointegrator 1 complex subunit
3; K01529 [EC:3.6.1.-]
Length=2202
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 45/77 (58%), Gaps = 0/77 (0%)
Query 4 IQGRMWSTMTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLLHRL 63
I R+W +PLRQF +LP +L ++E+K L D+ D+ EIG ++ +G + +
Sbjct 1116 IDKRLWGWASPLRQFSILPPHILTRLEEKKLTVDKLKDMRKDEIGHILHHVNIGLKVKQC 1175
Query 64 IHSFPKLELAAFVQPLS 80
+H P + + A +QP++
Sbjct 1176 VHQIPSVMMEASIQPIT 1192
> dre:100332599 mutagen-sensitive 308-like
Length=864
Score = 58.2 bits (139), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 43/80 (53%), Gaps = 0/80 (0%)
Query 1 CNEIQGRMWSTMTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLL 60
C + R+W PLRQF LP L ++E K+L D+ D+ EIG ++ +G +
Sbjct 24 CKVMDKRLWGWAHPLRQFNTLPASALARMEDKNLTIDKLRDMGKDEIGHMLHHVNIGLKV 83
Query 61 HRLIHSFPKLELAAFVQPLS 80
+ +H P + L + +QP++
Sbjct 84 KQCVHQIPAILLESSIQPIT 103
> sce:YGR271W SLH1; Putative RNA helicase related to Ski2p, involved
in translation inhibition of non-poly(A) mRNAs; required
for repressing propagation of dsRNA viruses (EC:3.6.1.-);
K01529 [EC:3.6.1.-]
Length=1967
Score = 54.3 bits (129), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 27/80 (33%), Positives = 46/80 (57%), Gaps = 1/80 (1%)
Query 1 CNEIQGRMWSTMTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLL 60
C I+ R+W+ PL QF LPE ++R+I + +L + E+GELV K G L
Sbjct 930 CKSIEKRLWAFDHPLCQFD-LPENIIRRIRDTKPSMEHLLELEADELGELVHNKKAGSRL 988
Query 61 HRLIHSFPKLELAAFVQPLS 80
++++ FPK+ + A + P++
Sbjct 989 YKILSRFPKINIEAEIFPIT 1008
> ath:AT5G61140 DEAD box RNA helicase, putative; K01529 [EC:3.6.1.-]
Length=2146
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 40/86 (46%), Gaps = 22/86 (25%)
Query 1 CNEIQGRMWSTMTPLRQFKVLPEELLRKIEKKDLPFDR------YYDLTSTEIGELVRVP 54
C + ++W PLRQF ++DLP DR Y++ EIG L+R
Sbjct 1143 CKAVDRQLWPHQHPLRQF------------ERDLPSDRRDDLDHLYEMEEKEIGALIRYN 1190
Query 55 KMGKLLHRLIHSFPKLELAAFVQPLS 80
G R + FP ++LAA V P++
Sbjct 1191 PGG----RHLGYFPSIQLAATVSPIT 1212
> mmu:446101 Xrra1, AI449753; X-ray radiation resistance associated
1
Length=786
Score = 29.3 bits (64), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 34/60 (56%), Gaps = 4/60 (6%)
Query 3 EIQGRMWSTMTPLRQ--FKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLL 60
E+Q +MW TP Q + VLP + + +++ ++ F Y +++E ++ +P M ++L
Sbjct 298 ELQPQMWIFETPDEQPNYTVLP--MKKDVDRTEVVFSSYPGFSTSETAKVCSLPPMFEIL 355
> mmu:319880 Tmcc3, AW488095, C630016B22Rik, C88213, MGC30586,
Tmcc1; transmembrane and coiled coil domains 3
Length=446
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 19/60 (31%), Positives = 27/60 (45%), Gaps = 1/60 (1%)
Query 5 QGRMWSTMTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIG-ELVRVPKMGKLLHRL 63
QG++ M LR+ KV +L IE + F R Y S + E R ++ LH L
Sbjct 253 QGKIAKIMEELREIKVTQTQLAEDIEALKVQFKREYGFISQTLQEERYRYERLEDQLHDL 312
> dre:792182 eef2a.1, MGC113191, si:dkey-110c1.3, zgc:113191;
eukaryotic translation elongation factor 2a, tandem duplicate
1; K03234 elongation factor 2
Length=854
Score = 28.1 bits (61), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 23/46 (50%), Gaps = 2/46 (4%)
Query 22 PEELLRKIEK--KDLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIH 65
PEE ++K+E K L D Y+D T+ + E P KL +H
Sbjct 244 PEEYIKKVEDMIKRLWGDSYFDSTTGKFSESATSPDGKKLPRTFVH 289
> tgo:TGME49_103850 hypothetical protein
Length=203
Score = 28.1 bits (61), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 18/65 (27%), Positives = 35/65 (53%), Gaps = 0/65 (0%)
Query 11 TMTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKL 70
TM+ L + + ++ L K E ++P+D+Y+ + + ++ K GK+++ L S
Sbjct 93 TMSTLVEAGFMTKKELEKFESFEIPYDKYWLPITWSMTHVLDARKSGKVINDLEMSKLVD 152
Query 71 ELAAF 75
EL AF
Sbjct 153 ELRAF 157
Lambda K H
0.324 0.141 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2058492688
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40