bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2811_orf1
Length=90
Score E
Sequences producing significant alignments: (Bits) Value
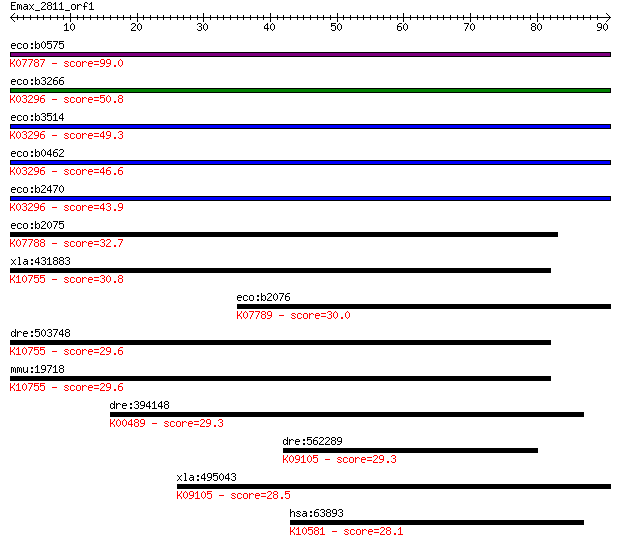
eco:b0575 cusA, ECK0567, JW0564, silA, ybdE; copper/silver eff... 99.0 3e-21
eco:b3266 acrF, ECK3253, envD, JW3234; multidrug efflux system... 50.8 1e-06
eco:b3514 mdtF, ECK3498, JW3482, yhiV; multidrug transporter, ... 49.3 3e-06
eco:b0462 acrB, acrE, ECK0456, JW0451; multidrug efflux system... 46.6 2e-05
eco:b2470 acrD, ECK2465, JW2454, yffA; aminoglycoside/multidru... 43.9 1e-04
eco:b2075 mdtB, ECK2071, JW2060, yegN; multidrug efflux system... 32.7 0.30
xla:431883 rfc2, MGC81391, rfc40; replication factor C (activa... 30.8 1.0
eco:b2076 mdtC, ECK2072, JW2061, yegO; multidrug efflux system... 30.0 1.9
dre:503748 rfc2, zgc:110810; replication factor C (activator 1... 29.6 2.2
mmu:19718 Rfc2, 2610008M13Rik, 40kDa, AI326953, MGC117486, Rec... 29.6 2.6
dre:394148 cyp7a1a, MGC63920, cyp7a1, zgc:63920; cytochrome P4... 29.3 3.0
dre:562289 tfeb; transcription factor EB; K09105 transcription... 29.3 3.4
xla:495043 tfe3; transcription factor binding to IGHM enhancer... 28.5 4.9
hsa:63893 UBE2O, E2-230K, FLJ12878, KIAA1734; ubiquitin-conjug... 28.1 6.9
> eco:b0575 cusA, ECK0567, JW0564, silA, ybdE; copper/silver efflux
system, membrane component; K07787 Cu(I)/Ag(I) efflux
system membrane protein CusA
Length=1047
Score = 99.0 bits (245), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 52/90 (57%), Positives = 67/90 (74%), Gaps = 0/90 (0%)
Query 1 VARVELGPDERRGIAELNGEGEVASGIVLQRFGMNALDVIENVKKRFKEIATSLPKSVEI 60
VA+V++GP+ RRGIAELNGEGEVA G+V+ R G NA +VI VK + + + +SLP+ VEI
Sbjct 262 VAKVQIGPEMRRGIAELNGEGEVAGGVVILRSGKNAREVIAAVKDKLETLKSSLPEGVEI 321
Query 61 VPVYDRSNLIYAAIETLKRTLLEESVVVAL 90
V YDRS LI AI+ L LLEE +VVA+
Sbjct 322 VTTYDRSQLIDRAIDNLSGKLLEEFIVVAV 351
> eco:b3266 acrF, ECK3253, envD, JW3234; multidrug efflux system
protein; K03296 hydrophobic/amphiphilic exporter-1 (mainly
G- bacteria), HAE1 family
Length=1034
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 49/90 (54%), Gaps = 1/90 (1%)
Query 1 VARVELGPDERRGIAELNGEGEVASGIVLQRFGMNALDVIENVKKRFKEIATSLPKSVEI 60
VARVELG + IA +NG+ GI L G NALD + +K + E+ P+ +++
Sbjct 265 VARVELGGENYNVIARINGKPAAGLGIKLAT-GANALDTAKAIKAKLAELQPFFPQGMKV 323
Query 61 VPVYDRSNLIYAAIETLKRTLLEESVVVAL 90
+ YD + + +I + +TL E ++V L
Sbjct 324 LYPYDTTPFVQLSIHEVVKTLFEAIMLVFL 353
> eco:b3514 mdtF, ECK3498, JW3482, yhiV; multidrug transporter,
RpoS-dependent; K03296 hydrophobic/amphiphilic exporter-1
(mainly G- bacteria), HAE1 family
Length=1037
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 32/91 (35%), Positives = 52/91 (57%), Gaps = 3/91 (3%)
Query 1 VARVELGPDERRGIAELNGEGEVASGIVLQ-RFGMNALDVIENVKKRFKEIATSLPKSVE 59
VARVELG ++ +A NG+ A+GI ++ G NALD VK+ ++ P S++
Sbjct 265 VARVELGAEDYSTVARYNGKP--AAGIAIKLAAGANALDTSRAVKEELNRLSAYFPASLK 322
Query 60 IVPVYDRSNLIYAAIETLKRTLLEESVVVAL 90
V YD + I +I+ + +TL+E ++V L
Sbjct 323 TVYPYDTTPFIEISIQEVFKTLVEAIILVFL 353
> eco:b0462 acrB, acrE, ECK0456, JW0451; multidrug efflux system
protein; K03296 hydrophobic/amphiphilic exporter-1 (mainly
G- bacteria), HAE1 family
Length=1049
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 47/90 (52%), Gaps = 1/90 (1%)
Query 1 VARVELGPDERRGIAELNGEGEVASGIVLQRFGMNALDVIENVKKRFKEIATSLPKSVEI 60
VA++ELG + IAE NG+ GI L G NALD ++ ++ P ++I
Sbjct 265 VAKIELGGENYDIIAEFNGQPASGLGIKLAT-GANALDTAAAIRAELAKMEPFFPSGLKI 323
Query 61 VPVYDRSNLIYAAIETLKRTLLEESVVVAL 90
V YD + + +I + +TL+E ++V L
Sbjct 324 VYPYDTTPFVKISIHEVVKTLVEAIILVFL 353
> eco:b2470 acrD, ECK2465, JW2454, yffA; aminoglycoside/multidrug
efflux system; K03296 hydrophobic/amphiphilic exporter-1
(mainly G- bacteria), HAE1 family
Length=1037
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 28/90 (31%), Positives = 46/90 (51%), Gaps = 1/90 (1%)
Query 1 VARVELGPDERRGIAELNGEGEVASGIVLQRFGMNALDVIENVKKRFKEIATSLPKSVEI 60
VA VE+G ++ ++ NG+ G+ L G N + E V R E+A P +E
Sbjct 265 VATVEMGAEKYDYLSRFNGKPASGLGVKLAS-GANEMATAELVLNRLDELAQYFPHGLEY 323
Query 61 VPVYDRSNLIYAAIETLKRTLLEESVVVAL 90
Y+ ++ + A+IE + +TLLE +V L
Sbjct 324 KVAYETTSFVKASIEDVVKTLLEAIALVFL 353
> eco:b2075 mdtB, ECK2071, JW2060, yegN; multidrug efflux system,
subunit B; K07788 RND superfamily, multidrug transport protein
MdtB
Length=1040
Score = 32.7 bits (73), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 22/82 (26%), Positives = 42/82 (51%), Gaps = 1/82 (1%)
Query 1 VARVELGPDERRGIAELNGEGEVASGIVLQRFGMNALDVIENVKKRFKEIATSLPKSVEI 60
VA VE G + A N E + + Q G N + +++++ ++ SLPKSV++
Sbjct 268 VATVEQGAENSWLGAWANKEQAIVMNVQRQP-GANIISTADSIRQMLPQLTESLPKSVKV 326
Query 61 VPVYDRSNLIYAAIETLKRTLL 82
+ DR+ I A+++ + L+
Sbjct 327 TVLSDRTTNIRASVDDTQFELM 348
> xla:431883 rfc2, MGC81391, rfc40; replication factor C (activator
1) 2, 40kDa; K10755 replication factor C subunit 2/4
Length=348
Score = 30.8 bits (68), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 21/82 (25%), Positives = 37/82 (45%), Gaps = 13/82 (15%)
Query 1 VARVELGPDERRGIAELNGEGEVASGIVLQRFGMNALDVIENVKKRFKEIATSLPKSVEI 60
+AR LGP + + ELN + +DV+ N K F + +LPK
Sbjct 84 LARALLGPTMKDAVLELNASND------------RGIDVVRNKIKMFAQQKVTLPKGRHK 131
Query 61 VPVYDRSN-LIYAAIETLKRTL 81
+ + D ++ + A + L+RT+
Sbjct 132 IIILDEADSMTDGAQQALRRTM 153
> eco:b2076 mdtC, ECK2072, JW2061, yegO; multidrug efflux system,
subunit C; K07789 RND superfamily, multidrug transport protein
MdtC
Length=1025
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 13/56 (23%), Positives = 34/56 (60%), Gaps = 0/56 (0%)
Query 35 NALDVIENVKKRFKEIATSLPKSVEIVPVYDRSNLIYAAIETLKRTLLEESVVVAL 90
N + +++++ + E+ ++P ++++ DRS I A++E +++TL+ +V L
Sbjct 292 NIIQTVDSIRAKLPELQETIPAAIDLQIAQDRSPTIRASLEEVEQTLIISVALVIL 347
> dre:503748 rfc2, zgc:110810; replication factor C (activator
1) 2; K10755 replication factor C subunit 2/4
Length=349
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 21/82 (25%), Positives = 37/82 (45%), Gaps = 13/82 (15%)
Query 1 VARVELGPDERRGIAELNGEGEVASGIVLQRFGMNALDVIENVKKRFKEIATSLPKSVEI 60
+AR LGP + + ELN + +DV+ N K F + +LPK
Sbjct 84 LARALLGPAMKDAVLELNASND------------RGIDVVRNKIKMFAQQKVTLPKGRHK 131
Query 61 VPVYDRSN-LIYAAIETLKRTL 81
+ + D ++ + A + L+RT+
Sbjct 132 IIILDEADSMTDGAQQALRRTM 153
> mmu:19718 Rfc2, 2610008M13Rik, 40kDa, AI326953, MGC117486, Recc2;
replication factor C (activator 1) 2; K10755 replication
factor C subunit 2/4
Length=349
Score = 29.6 bits (65), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 21/82 (25%), Positives = 37/82 (45%), Gaps = 13/82 (15%)
Query 1 VARVELGPDERRGIAELNGEGEVASGIVLQRFGMNALDVIENVKKRFKEIATSLPKSVEI 60
+AR LGP + + ELN + +DV+ N K F + +LPK
Sbjct 84 LARALLGPALKDAVLELNASND------------RGIDVVRNKIKMFAQQKVTLPKGRHK 131
Query 61 VPVYDRSN-LIYAAIETLKRTL 81
+ + D ++ + A + L+RT+
Sbjct 132 IIILDEADSMTDGAQQALRRTM 153
> dre:394148 cyp7a1a, MGC63920, cyp7a1, zgc:63920; cytochrome
P450, family 7, subfamily A, polypeptide 1a (EC:1.14.13.17);
K00489 cytochrome P450, family 7, subfamily A (cholesterol
7alpha-monooxygenase) [EC:1.14.13.17]
Length=512
Score = 29.3 bits (64), Expect = 3.0, Method: Composition-based stats.
Identities = 20/71 (28%), Positives = 35/71 (49%), Gaps = 8/71 (11%)
Query 16 ELNGEGEVASGIVLQRFGMNALDVIENVKKRFKEIATSLPKSVEIVPVYDRSNLIYAAIE 75
EL+G+ +A + +NALD + K F + LP + V+ + Y+A E
Sbjct 192 ELDGDQSIARQQAQKALVLNALDNFKEFDKIFPALIAGLP-----IHVFKSA---YSARE 243
Query 76 TLKRTLLEESV 86
L +T+L E++
Sbjct 244 KLAKTMLHENL 254
> dre:562289 tfeb; transcription factor EB; K09105 transcription
factor E3-related
Length=493
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 10/38 (26%), Positives = 25/38 (65%), Gaps = 0/38 (0%)
Query 42 NVKKRFKEIATSLPKSVEIVPVYDRSNLIYAAIETLKR 79
N+ R KE+ T +PK+ ++ +++ ++ A+++ +KR
Sbjct 260 NINDRIKELGTMIPKTNDLDVRWNKGTILRASVDYIKR 297
> xla:495043 tfe3; transcription factor binding to IGHM enhancer
3; K09105 transcription factor E3-related
Length=429
Score = 28.5 bits (62), Expect = 4.9, Method: Composition-based stats.
Identities = 17/70 (24%), Positives = 38/70 (54%), Gaps = 5/70 (7%)
Query 26 GIVLQRFGMNALDVIE-----NVKKRFKEIATSLPKSVEIVPVYDRSNLIYAAIETLKRT 80
I+ +R ++ ++IE N+ R KE+ T +PKS + +++ ++ A++E +++
Sbjct 219 AIIKERQKKDSHNLIERRRRFNINDRIKELGTLIPKSSDPEVRWNKGTILKASVEYIRKL 278
Query 81 LLEESVVVAL 90
E+ V L
Sbjct 279 QKEQQRVREL 288
> hsa:63893 UBE2O, E2-230K, FLJ12878, KIAA1734; ubiquitin-conjugating
enzyme E2O (EC:6.3.2.19); K10581 ubiquitin-conjugating
enzyme E2 O [EC:6.3.2.19]
Length=1292
Score = 28.1 bits (61), Expect = 6.9, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 24/44 (54%), Gaps = 0/44 (0%)
Query 43 VKKRFKEIATSLPKSVEIVPVYDRSNLIYAAIETLKRTLLEESV 86
V+K +ATSLP+ + + DR +L A I+ RT E+ +
Sbjct 958 VRKEMALLATSLPEGIMVKTFEDRMDLFSALIKGPTRTPYEDGL 1001
Lambda K H
0.316 0.136 0.355
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2012433808
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40