bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2748_orf1
Length=117
Score E
Sequences producing significant alignments: (Bits) Value
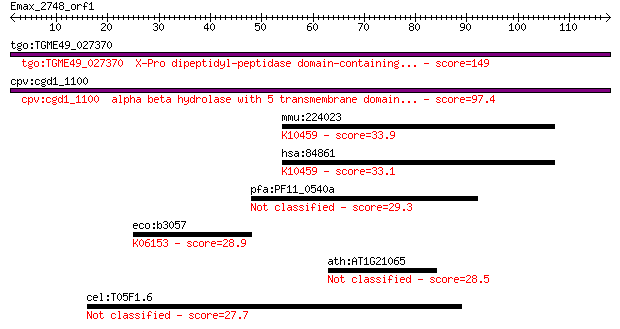
tgo:TGME49_027370 X-Pro dipeptidyl-peptidase domain-containing... 149 1e-36
cpv:cgd1_1100 alpha beta hydrolase with 5 transmembrane domain... 97.4 1e-20
mmu:224023 Klhl22, 2610318I18Rik, Kelchl; kelch-like 22 (Droso... 33.9 0.12
hsa:84861 KLHL22, KELCHL; kelch-like 22 (Drosophila); K10459 k... 33.1 0.20
pfa:PF11_0540a conserved Plasmodium protein 29.3 2.8
eco:b3057 bacA, ECK3047, JW3029, uppP; undecaprenyl pyrophosph... 28.9 4.4
ath:AT1G21065 hypothetical protein 28.5 5.2
cel:T05F1.6 hsr-9; hypothetical protein 27.7 9.8
> tgo:TGME49_027370 X-Pro dipeptidyl-peptidase domain-containing
protein (EC:3.4.14.11); K06978
Length=1177
Score = 149 bits (377), Expect = 1e-36, Method: Composition-based stats.
Identities = 65/117 (55%), Positives = 84/117 (71%), Gaps = 0/117 (0%)
Query 1 GKVAGGGLSYDGMLGLSLASRGGIDTIISLFTPINIMNELVAPGGLLCYSFLHDYTQLTN 60
GKV GGG+SYDGM GLS A+ GG+D ++SLFTP+++ +L+ PGG +C SFL DY +T
Sbjct 325 GKVGGGGISYDGMTGLSTAAAGGVDAVLSLFTPMHVFGDLLVPGGFVCSSFLKDYAGMTY 384
Query 61 NFEQNGSPWRHMLNSPLQFPFHVLLGFLFSFGGASAVLGYEEDLHKAINSHKNNWRM 117
FE+ G+P HMLN+P ++P HVLLGF +FG S V G E DL +AI HK NW M
Sbjct 385 GFERAGTPLAHMLNNPWKYPLHVLLGFSVAFGPGSGVFGREGDLARAIMGHKKNWDM 441
> cpv:cgd1_1100 alpha beta hydrolase with 5 transmembrane domains
; K06978
Length=1103
Score = 97.4 bits (241), Expect = 1e-20, Method: Composition-based stats.
Identities = 47/134 (35%), Positives = 70/134 (52%), Gaps = 17/134 (12%)
Query 1 GKVAGGGLSYDGMLGLSLAS-----------------RGGIDTIISLFTPINIMNELVAP 43
GKV GG+SYDGM + A+ +D + +L +P+N++ EL+ P
Sbjct 518 GKVGVGGISYDGMAAIKTAALSGNNESESSSEGFRSDNNLVDAVFALSSPMNVIKELIEP 577
Query 44 GGLLCYSFLHDYTQLTNNFEQNGSPWRHMLNSPLQFPFHVLLGFLFSFGGASAVLGYEED 103
GGL+C + DY +T +FEQ GSP H L + +PF +++ FL G S V GY
Sbjct 578 GGLICKPLVEDYYSITYSFEQYGSPLLHFLKTVAYYPFKLVVAFLLVIGNVSPVQGYSSI 637
Query 104 LHKAINSHKNNWRM 117
+A+ H+ NW M
Sbjct 638 KKQALEMHQKNWDM 651
> mmu:224023 Klhl22, 2610318I18Rik, Kelchl; kelch-like 22 (Drosophila);
K10459 kelch-like protein 22
Length=634
Score = 33.9 bits (76), Expect = 0.12, Method: Composition-based stats.
Identities = 16/56 (28%), Positives = 30/56 (53%), Gaps = 3/56 (5%)
Query 54 DYTQLTNNFEQNGSPWRHMLNSPLQFPFH---VLLGFLFSFGGASAVLGYEEDLHK 106
DY + T+ ++ + W + + P++ +H LL LF GG++ GY D+H+
Sbjct 458 DYLKETHCYDPGSNTWHTLADGPVRRAWHGMAALLDKLFVIGGSNNDAGYRRDVHQ 513
> hsa:84861 KLHL22, KELCHL; kelch-like 22 (Drosophila); K10459
kelch-like protein 22
Length=634
Score = 33.1 bits (74), Expect = 0.20, Method: Composition-based stats.
Identities = 15/56 (26%), Positives = 30/56 (53%), Gaps = 3/56 (5%)
Query 54 DYTQLTNNFEQNGSPWRHMLNSPLQFPFH---VLLGFLFSFGGASAVLGYEEDLHK 106
DY + T+ ++ + W + + P++ +H LL L+ GG++ GY D+H+
Sbjct 458 DYLKETHCYDPGSNTWHTLADGPVRRAWHGMATLLNKLYVIGGSNNDAGYRRDVHQ 513
> pfa:PF11_0540a conserved Plasmodium protein
Length=1467
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 14/46 (30%), Positives = 23/46 (50%), Gaps = 2/46 (4%)
Query 48 CYSFLHDYTQLTNNFEQNGSP--WRHMLNSPLQFPFHVLLGFLFSF 91
CY FL+ Y + N+ N SP W + S + +L+ F +S+
Sbjct 330 CYLFLNTYNENLNDITMNTSPSRWNKIKLSSWGYKGSLLINFFYSY 375
> eco:b3057 bacA, ECK3047, JW3029, uppP; undecaprenyl pyrophosphate
phosphatase (EC:3.6.1.27); K06153 undecaprenyl-diphosphatase
[EC:3.6.1.27]
Length=273
Score = 28.9 bits (63), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 15/23 (65%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 25 DTIISLFTPINIMNELVAPGGLL 47
DTI SLF PIN+M LV G LL
Sbjct 111 DTIKSLFNPINVMYALVVGGLLL 133
> ath:AT1G21065 hypothetical protein
Length=217
Score = 28.5 bits (62), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 9/21 (42%), Positives = 12/21 (57%), Gaps = 0/21 (0%)
Query 63 EQNGSPWRHMLNSPLQFPFHV 83
E N +PWRH + P P H+
Sbjct 150 EGNSAPWRHTMEGPDDMPAHI 170
> cel:T05F1.6 hsr-9; hypothetical protein
Length=1165
Score = 27.7 bits (60), Expect = 9.8, Method: Composition-based stats.
Identities = 19/73 (26%), Positives = 35/73 (47%), Gaps = 8/73 (10%)
Query 16 LSLASRGGIDTIISLFTPINIMNELVAPGGLLCYSFLHDYTQLTNNFEQNGSPWRHMLNS 75
L+ A+R ++ S+F N+MN + GG++ T+ N+F++ S + +L S
Sbjct 935 LTSANRSNSASVPSMFKKKNLMNFITQNGGIV--------TEQLNSFQERYSNYEPLLIS 986
Query 76 PLQFPFHVLLGFL 88
+ H L L
Sbjct 987 DTYYRTHKYLAAL 999
Lambda K H
0.322 0.141 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2032807080
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40