bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2728_orf2
Length=153
Score E
Sequences producing significant alignments: (Bits) Value
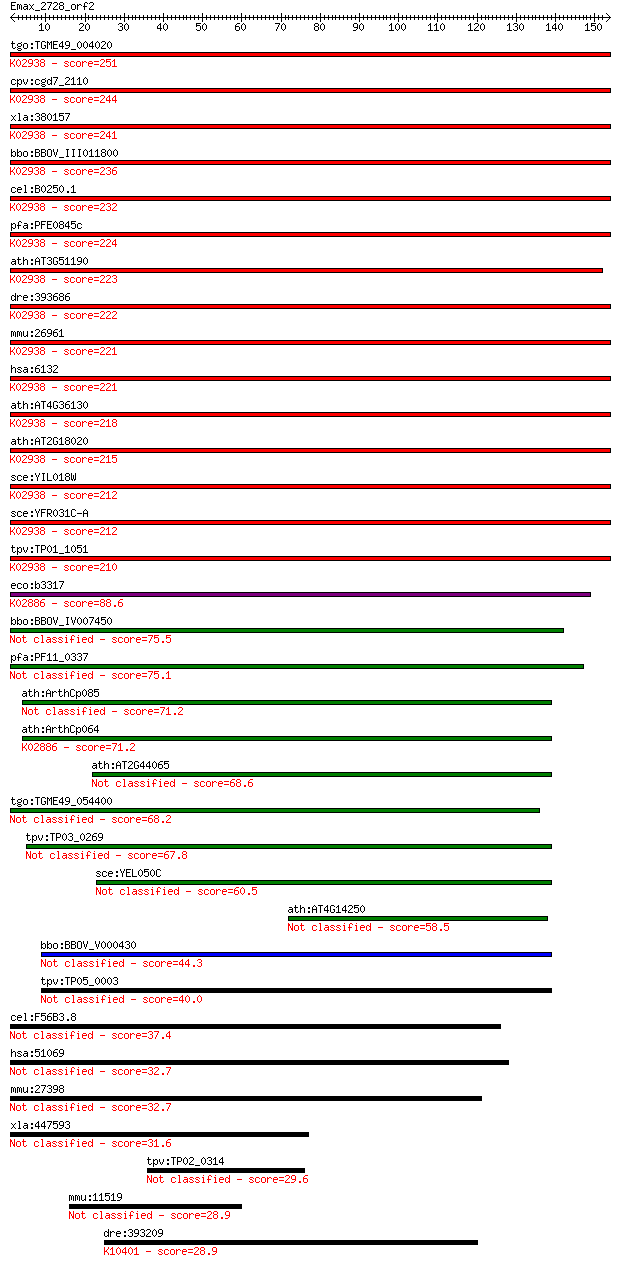
tgo:TGME49_004020 60S ribosomal protein L8, putative ; K02938 ... 251 5e-67
cpv:cgd7_2110 60S ribosomal proteins L8/L2 ; K02938 large subu... 244 9e-65
xla:380157 rpl8, MGC53203; ribosomal protein L8; K02938 large ... 241 6e-64
bbo:BBOV_III011800 17.m08006; ribosomal protein L2 family prot... 236 2e-62
cel:B0250.1 rpl-2; Ribosomal Protein, Large subunit family mem... 232 4e-61
pfa:PFE0845c 60S ribosomal protein L8, putative; K02938 large ... 224 8e-59
ath:AT3G51190 structural constituent of ribosome; K02938 large... 223 1e-58
dre:393686 rpl8, MGC73105, zgc:73105, zgc:77641; ribosomal pro... 222 3e-58
mmu:26961 Rpl8; ribosomal protein L8; K02938 large subunit rib... 221 7e-58
hsa:6132 RPL8; ribosomal protein L8; K02938 large subunit ribo... 221 7e-58
ath:AT4G36130 60S ribosomal protein L8 (RPL8C); K02938 large s... 218 6e-57
ath:AT2G18020 EMB2296 (embryo defective 2296); structural cons... 215 4e-56
sce:YIL018W RPL2B, LOT2, RPL5A; Rpl2bp; K02938 large subunit r... 212 3e-55
sce:YFR031C-A RPL2A, RPL5B; Rpl2ap; K02938 large subunit ribos... 212 3e-55
tpv:TP01_1051 60S ribosomal protein L8; K02938 large subunit r... 210 2e-54
eco:b3317 rplB, ECK3304, JW3279; 50S ribosomal subunit protein... 88.6 6e-18
bbo:BBOV_IV007450 23.m05961; ribosomal protein L2 75.5 7e-14
pfa:PF11_0337 mitochondrial ribosomal protein L2 precursor 75.1 8e-14
ath:ArthCp085 rpl2; 50S ribosomal protein L2 71.2 1e-12
ath:ArthCp064 rpl2; 50S ribosomal protein L2; K02886 large sub... 71.2 1e-12
ath:AT2G44065 ribosomal protein L2 family protein 68.6 9e-12
tgo:TGME49_054400 50S ribosomal protein l2, putative 68.2 1e-11
tpv:TP03_0269 60S ribosomal protein L2 67.8 1e-11
sce:YEL050C RML2; Mitochondrial ribosomal protein of the large... 60.5 2e-09
ath:AT4G14250 structural constituent of ribosome 58.5 8e-09
bbo:BBOV_V000430 ribosomal protein L2 44.3 1e-04
tpv:TP05_0003 rpl2; ribosomal protein L2 40.0 0.003
cel:F56B3.8 hypothetical protein 37.4 0.018
hsa:51069 MRPL2, MRP-L14, RPML14; mitochondrial ribosomal prot... 32.7 0.42
mmu:27398 Mrpl2, CGI-22, MRP-L14, Rpml14; mitochondrial riboso... 32.7 0.45
xla:447593 MGC84466 protein 31.6 1.2
tpv:TP02_0314 hypothetical protein 29.6 4.0
mmu:11519 Add2, 2900072M03Rik; adducin 2 (beta) 28.9 6.3
dre:393209 MGC55995; zgc:55995; K10401 kinesin family member 1... 28.9 7.3
> tgo:TGME49_004020 60S ribosomal protein L8, putative ; K02938
large subunit ribosomal protein L8e
Length=260
Score = 251 bits (642), Expect = 5e-67, Method: Compositional matrix adjust.
Identities = 116/153 (75%), Positives = 138/153 (90%), Gaps = 0/153 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+AVEG++TGQFI+CGK AAL+IGN+LPL K+PEG+VV +VEEK GDRG LA+TSG++AT+
Sbjct 77 VAVEGMYTGQFIYCGKNAALTIGNILPLNKMPEGTVVSNVEEKAGDRGTLARTSGTYATI 136
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
VGHS++ K+R+RLPSGARKT+ SR ++G+VAGGGRIDKP+LKAG AYHKYK KRNCW
Sbjct 137 VGHSDDGSKTRIRLPSGARKTVSGYSRGMVGIVAGGGRIDKPMLKAGNAYHKYKVKRNCW 196
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRHA 153
KVRGVAMNPVEHPHGGGNHQHIGHPSTVSR A
Sbjct 197 PKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRMA 229
> cpv:cgd7_2110 60S ribosomal proteins L8/L2 ; K02938 large subunit
ribosomal protein L8e
Length=261
Score = 244 bits (623), Expect = 9e-65, Method: Compositional matrix adjust.
Identities = 111/153 (72%), Positives = 134/153 (87%), Gaps = 0/153 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+AVEG+HTGQF++CGK A LS+GNVLP+G++PEG+++ S EEK GDRG LA+TSG++A +
Sbjct 77 IAVEGLHTGQFVYCGKNATLSVGNVLPVGQIPEGTIISSCEEKAGDRGKLARTSGTYAII 136
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
VG SE+ K+R+RLPSGARKT+ S SR IIG+VA GGRIDKP+LKAG YHKYK KRNCW
Sbjct 137 VGQSEDGTKTRIRLPSGARKTISSQSRAIIGIVAAGGRIDKPVLKAGNNYHKYKVKRNCW 196
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRHA 153
KVRGV+MNPV+HPHGGGNHQHIGHPSTVSR A
Sbjct 197 PKVRGVSMNPVDHPHGGGNHQHIGHPSTVSRSA 229
> xla:380157 rpl8, MGC53203; ribosomal protein L8; K02938 large
subunit ribosomal protein L8e
Length=257
Score = 241 bits (615), Expect = 6e-64, Method: Compositional matrix adjust.
Identities = 110/153 (71%), Positives = 134/153 (87%), Gaps = 0/153 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+A EGIHTGQF++CGKKA L+IGNVLP+G +PEG++VC VEEKPGDRG LA+ SG++ATV
Sbjct 77 VAAEGIHTGQFVYCGKKAQLNIGNVLPVGTMPEGTIVCCVEEKPGDRGKLARASGNYATV 136
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
+ H+ ET K+RV+LPSG++K + S +R I+G+VAGGGRIDKP+LKAG AYHKYKAKRNCW
Sbjct 137 ISHNPETKKTRVKLPSGSKKVISSANRAIVGVVAGGGRIDKPILKAGRAYHKYKAKRNCW 196
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRHA 153
+VRGVAMNPVEHP GGGNHQHIG PST+ R A
Sbjct 197 PRVRGVAMNPVEHPFGGGNHQHIGKPSTIRRDA 229
> bbo:BBOV_III011800 17.m08006; ribosomal protein L2 family protein;
K02938 large subunit ribosomal protein L8e
Length=257
Score = 236 bits (602), Expect = 2e-62, Method: Compositional matrix adjust.
Identities = 109/153 (71%), Positives = 136/153 (88%), Gaps = 0/153 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+AVEG++TGQ+++ G+++ L+IGN +P+GK+PEG+V+ S+EEK GDRG LAK SG++AT+
Sbjct 77 VAVEGMYTGQYVYFGRESRLNIGNCMPVGKMPEGTVISSLEEKRGDRGRLAKASGTYATI 136
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
+GHS++ +RVRLPSGARK++ SNSR IIGLVAGGGRIDKPLLKAGTAY KYKAKRNCW
Sbjct 137 IGHSDDGKMTRVRLPSGARKSVESNSRAIIGLVAGGGRIDKPLLKAGTAYFKYKAKRNCW 196
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRHA 153
+VRGV MNPV+HPHGGGNHQHIG PSTVSR A
Sbjct 197 PRVRGVTMNPVDHPHGGGNHQHIGKPSTVSRGA 229
> cel:B0250.1 rpl-2; Ribosomal Protein, Large subunit family member
(rpl-2); K02938 large subunit ribosomal protein L8e
Length=260
Score = 232 bits (591), Expect = 4e-61, Method: Compositional matrix adjust.
Identities = 105/153 (68%), Positives = 130/153 (84%), Gaps = 0/153 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+A EG+HTGQFIHCG KA + IGN++P+G +PEG+ +C+VE K GDRG +A+ SG++ATV
Sbjct 77 VAAEGMHTGQFIHCGAKAQIQIGNIVPVGTLPEGTTICNVENKSGDRGVIARASGNYATV 136
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
+ H+ +T K+R+RLPSGA+K + S +R +IGLVAGGGR DKPLLKAG +YHKYKAKRN W
Sbjct 137 IAHNPDTKKTRIRLPSGAKKVVQSVNRAMIGLVAGGGRTDKPLLKAGRSYHKYKAKRNSW 196
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRHA 153
+VRGVAMNPVEHPHGGGNHQHIGHPSTV R A
Sbjct 197 PRVRGVAMNPVEHPHGGGNHQHIGHPSTVRRDA 229
> pfa:PFE0845c 60S ribosomal protein L8, putative; K02938 large
subunit ribosomal protein L8e
Length=260
Score = 224 bits (571), Expect = 8e-59, Method: Compositional matrix adjust.
Identities = 102/153 (66%), Positives = 130/153 (84%), Gaps = 0/153 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+A EG+ TGQ+I CG KA LS+GN+LP+GK+PEG+++C++E + G+RG L K SG +ATV
Sbjct 77 IASEGMFTGQYISCGTKAPLSVGNILPIGKMPEGTLICNLEHRTGNRGTLVKASGCYATV 136
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
VG SE+ K++VRLPSGA+KT+ + +R ++G+V GGRIDKP+LKAG A+HKY+ KRNCW
Sbjct 137 VGQSEDGKKTKVRLPSGAKKTIDAKARAMVGVVGAGGRIDKPILKAGVAHHKYRVKRNCW 196
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRHA 153
KVRGVAMNPVEHPHGGGNHQHIGHPSTVSR A
Sbjct 197 PKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRSA 229
> ath:AT3G51190 structural constituent of ribosome; K02938 large
subunit ribosomal protein L8e
Length=260
Score = 223 bits (569), Expect = 1e-58, Method: Compositional matrix adjust.
Identities = 104/151 (68%), Positives = 126/151 (83%), Gaps = 0/151 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+A EG++TGQ+++CGKKA L +GNVLPLG +PEG+V+C+VE GDRGALA+ SG +A V
Sbjct 78 VAAEGMYTGQYLYCGKKANLMVGNVLPLGSIPEGAVICNVELHVGDRGALARASGDYAIV 137
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
+ H+ E+ +RV+LPSG++K LPS R +IG VAGGGR +KPLLKAG AYHKYKAKRNCW
Sbjct 138 IAHNPESNTTRVKLPSGSKKILPSACRAMIGQVAGGGRTEKPLLKAGNAYHKYKAKRNCW 197
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSR 151
VRGVAMNPVEHPHGGGNHQHIGH STV R
Sbjct 198 PVVRGVAMNPVEHPHGGGNHQHIGHASTVRR 228
> dre:393686 rpl8, MGC73105, zgc:73105, zgc:77641; ribosomal protein
L8; K02938 large subunit ribosomal protein L8e
Length=257
Score = 222 bits (566), Expect = 3e-58, Method: Compositional matrix adjust.
Identities = 110/153 (71%), Positives = 134/153 (87%), Gaps = 0/153 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+A EGIHTGQFI+CGKKA L+IGNVLP+G +PEG++VC +EEKPGDRG LA+ SG++ATV
Sbjct 77 IAAEGIHTGQFIYCGKKAQLNIGNVLPVGTMPEGTIVCCLEEKPGDRGKLARASGNYATV 136
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
+ H+ ET KSRV+LPSG++K + S +R ++G+VAGGGRIDKP+LKAG AYHKYKAKRNCW
Sbjct 137 ISHNPETKKSRVKLPSGSKKVISSANRAVVGVVAGGGRIDKPILKAGRAYHKYKAKRNCW 196
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRHA 153
+VRGVAMNPVEHP GGGNHQHIG PST+ R A
Sbjct 197 PRVRGVAMNPVEHPFGGGNHQHIGKPSTIRRDA 229
> mmu:26961 Rpl8; ribosomal protein L8; K02938 large subunit ribosomal
protein L8e
Length=257
Score = 221 bits (563), Expect = 7e-58, Method: Compositional matrix adjust.
Identities = 108/153 (70%), Positives = 134/153 (87%), Gaps = 0/153 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+A EGIHTGQF++CGKKA L+IGNVLP+G +PEG++VC +EEKPGDRG LA+ SG++ATV
Sbjct 77 IAAEGIHTGQFVYCGKKAQLNIGNVLPVGTMPEGTIVCCLEEKPGDRGKLARASGNYATV 136
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
+ H+ ET K+RV+LPSG++K + S +R ++G+VAGGGRIDKP+LKAG AYHKYKAKRNCW
Sbjct 137 ISHNPETKKTRVKLPSGSKKVISSANRAVVGVVAGGGRIDKPILKAGRAYHKYKAKRNCW 196
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRHA 153
+VRGVAMNPVEHP GGGNHQHIG PST+ R A
Sbjct 197 PRVRGVAMNPVEHPFGGGNHQHIGKPSTIRRDA 229
> hsa:6132 RPL8; ribosomal protein L8; K02938 large subunit ribosomal
protein L8e
Length=257
Score = 221 bits (563), Expect = 7e-58, Method: Compositional matrix adjust.
Identities = 108/153 (70%), Positives = 134/153 (87%), Gaps = 0/153 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+A EGIHTGQF++CGKKA L+IGNVLP+G +PEG++VC +EEKPGDRG LA+ SG++ATV
Sbjct 77 IAAEGIHTGQFVYCGKKAQLNIGNVLPVGTMPEGTIVCCLEEKPGDRGKLARASGNYATV 136
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
+ H+ ET K+RV+LPSG++K + S +R ++G+VAGGGRIDKP+LKAG AYHKYKAKRNCW
Sbjct 137 ISHNPETKKTRVKLPSGSKKVISSANRAVVGVVAGGGRIDKPILKAGRAYHKYKAKRNCW 196
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRHA 153
+VRGVAMNPVEHP GGGNHQHIG PST+ R A
Sbjct 197 PRVRGVAMNPVEHPFGGGNHQHIGKPSTIRRDA 229
> ath:AT4G36130 60S ribosomal protein L8 (RPL8C); K02938 large
subunit ribosomal protein L8e
Length=258
Score = 218 bits (555), Expect = 6e-57, Method: Compositional matrix adjust.
Identities = 99/153 (64%), Positives = 123/153 (80%), Gaps = 0/153 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+A EG++TGQF++CGKKA L +GNVLPL +PEG+V+C+VE GDRG A+ SG +A V
Sbjct 77 VAAEGMYTGQFLYCGKKATLVVGNVLPLRSIPEGAVICNVEHHVGDRGVFARASGDYAIV 136
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
+ H+ + SR++LPSG++K +PS R +IG VAGGGR +KP+LKAG AYHKY+ KRNCW
Sbjct 137 IAHNPDNDTSRIKLPSGSKKIVPSGCRAMIGQVAGGGRTEKPMLKAGNAYHKYRVKRNCW 196
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRHA 153
KVRGVAMNPVEHPHGGGNHQHIGH STV R A
Sbjct 197 PKVRGVAMNPVEHPHGGGNHQHIGHASTVRRDA 229
> ath:AT2G18020 EMB2296 (embryo defective 2296); structural constituent
of ribosome; K02938 large subunit ribosomal protein
L8e
Length=258
Score = 215 bits (547), Expect = 4e-56, Method: Compositional matrix adjust.
Identities = 99/153 (64%), Positives = 124/153 (81%), Gaps = 0/153 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+A EG++TGQF++CGKKA L +GNVLPL +PEG+VVC+VE GDRG LA+ SG +A V
Sbjct 77 VAAEGMYTGQFLYCGKKATLVVGNVLPLRSIPEGAVVCNVEHHVGDRGVLARASGDYAIV 136
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
+ H+ ++ +R++LPSG++K +PS R +IG VAGGGR +KP+LKAG AYHKY+ KRN W
Sbjct 137 IAHNPDSDTTRIKLPSGSKKIVPSGCRAMIGQVAGGGRTEKPMLKAGNAYHKYRVKRNSW 196
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRHA 153
KVRGVAMNPVEHPHGGGNHQHIGH STV R A
Sbjct 197 PKVRGVAMNPVEHPHGGGNHQHIGHASTVRRDA 229
> sce:YIL018W RPL2B, LOT2, RPL5A; Rpl2bp; K02938 large subunit
ribosomal protein L8e
Length=254
Score = 212 bits (540), Expect = 3e-55, Method: Compositional matrix adjust.
Identities = 103/153 (67%), Positives = 132/153 (86%), Gaps = 0/153 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+A EG+HTGQFI+ GKKA+L++GNVLPLG VPEG++V +VEEKPGDRGALA+ SG++ +
Sbjct 77 IANEGVHTGQFIYAGKKASLNVGNVLPLGSVPEGTIVSNVEEKPGDRGALARASGNYVII 136
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
+GH+ + K+RVRLPSGA+K + S++R +IG++AGGGR+DKPLLKAG A+HKY+ KRN W
Sbjct 137 IGHNPDENKTRVRLPSGAKKVISSDARGVIGVIAGGGRVDKPLLKAGRAFHKYRLKRNSW 196
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRHA 153
K RGVAMNPV+HPHGGGNHQHIG ST+SR A
Sbjct 197 PKTRGVAMNPVDHPHGGGNHQHIGKASTISRGA 229
> sce:YFR031C-A RPL2A, RPL5B; Rpl2ap; K02938 large subunit ribosomal
protein L8e
Length=254
Score = 212 bits (540), Expect = 3e-55, Method: Compositional matrix adjust.
Identities = 103/153 (67%), Positives = 132/153 (86%), Gaps = 0/153 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+A EG+HTGQFI+ GKKA+L++GNVLPLG VPEG++V +VEEKPGDRGALA+ SG++ +
Sbjct 77 IANEGVHTGQFIYAGKKASLNVGNVLPLGSVPEGTIVSNVEEKPGDRGALARASGNYVII 136
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
+GH+ + K+RVRLPSGA+K + S++R +IG++AGGGR+DKPLLKAG A+HKY+ KRN W
Sbjct 137 IGHNPDENKTRVRLPSGAKKVISSDARGVIGVIAGGGRVDKPLLKAGRAFHKYRLKRNSW 196
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRHA 153
K RGVAMNPV+HPHGGGNHQHIG ST+SR A
Sbjct 197 PKTRGVAMNPVDHPHGGGNHQHIGKASTISRGA 229
> tpv:TP01_1051 60S ribosomal protein L8; K02938 large subunit
ribosomal protein L8e
Length=258
Score = 210 bits (534), Expect = 2e-54, Method: Compositional matrix adjust.
Identities = 110/153 (71%), Positives = 134/153 (87%), Gaps = 0/153 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+AVEG+++GQF++ G A+L++GN LPLGK+PEG+VV +EEK GDRG +AKTSGS+ATV
Sbjct 77 VAVEGLYSGQFVYFGANASLAVGNCLPLGKMPEGTVVSCLEEKKGDRGRVAKTSGSYATV 136
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
+ HS++ +R+RLPSG RKT+ S +R I+G+VAGGGRIDKPLLKAG AYHKYKAKRNCW
Sbjct 137 ISHSDDGKTTRIRLPSGTRKTVSSAARGIVGVVAGGGRIDKPLLKAGVAYHKYKAKRNCW 196
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRHA 153
KVRGVAMNPVEHPHGGGNHQHIGHPST+SR A
Sbjct 197 PKVRGVAMNPVEHPHGGGNHQHIGHPSTISRLA 229
> eco:b3317 rplB, ECK3304, JW3279; 50S ribosomal subunit protein
L2; K02886 large subunit ribosomal protein L2
Length=273
Score = 88.6 bits (218), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 57/149 (38%), Positives = 82/149 (55%), Gaps = 8/149 (5%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
LA +G+ G I G AA+ GN LP+ +P GS V +VE KPG G LA+++G++ +
Sbjct 105 LAPKGLKAGDQIQSGVDAAIKPGNTLPMRNIPVGSTVHNVEMKPGKGGQLARSAGTYVQI 164
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
V + + +RL SG + + ++ R +G V + + L KAG A R
Sbjct 165 V--ARDGAYVTLRLRSGEMRKVEADCRATLGEVGNAEHMLRVLGKAGAARW-----RGVR 217
Query 121 HKVRGVAMNPVEHPHGGGNHQHIG-HPST 148
VRG AMNPV+HPHGGG ++ G HP T
Sbjct 218 PTVRGTAMNPVDHPHGGGEGRNFGKHPVT 246
> bbo:BBOV_IV007450 23.m05961; ribosomal protein L2
Length=332
Score = 75.5 bits (184), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 50/143 (34%), Positives = 73/143 (51%), Gaps = 11/143 (7%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
L G+ GQ + A + GN LPL +P S+V +VE +PG G +A+ G++ T+
Sbjct 150 LCPAGVRPGQILLASASAPIEPGNCLPLRHIPVNSIVHNVELRPGAGGQIARAGGTYVTI 209
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNC- 119
V E +R+ S + P + +G V+ + L KAGT +RN
Sbjct 210 VEKDEMFAT--LRMASTELRRFPLDCWATLGQVSNIEHNKRILKKAGT-------RRNLG 260
Query 120 WH-KVRGVAMNPVEHPHGGGNHQ 141
W VRG+AMNP HPHGGGN++
Sbjct 261 WRPSVRGIAMNPNAHPHGGGNNK 283
> pfa:PF11_0337 mitochondrial ribosomal protein L2 precursor
Length=333
Score = 75.1 bits (183), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 52/147 (35%), Positives = 78/147 (53%), Gaps = 8/147 (5%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
LA + G I K A ++ GN LPL +P GS++ ++E +PG G L + +G++AT+
Sbjct 153 LAPLLLRPGDKIIASKYANINPGNSLPLQNIPVGSIIHNIEMRPGAGGQLIRAAGTYATI 212
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
+ E ++L S + P IG V+ R + L KAG +++ KR
Sbjct 213 ISKDNEYAT--IKLKSTEIRKFPLQCWATIGQVSNLERHMRILGKAGV--NRWLGKRPV- 267
Query 121 HKVRGVAMNPVEHPHGGG-NHQHIGHP 146
VRGVAMNP +HPHGGG + +H P
Sbjct 268 --VRGVAMNPSKHPHGGGTSKKHTKRP 292
> ath:ArthCp085 rpl2; 50S ribosomal protein L2
Length=274
Score = 71.2 bits (173), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 47/139 (33%), Positives = 71/139 (51%), Gaps = 15/139 (10%)
Query 4 EGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATVVGH 63
G G I G + + +GN LPL +P G+ + ++E G G LA+ +G+ A ++
Sbjct 106 RGAIIGDTIVSGTEVPIKMGNALPLTDMPLGTAIHNIEITLGRGGQLARAAGAVAKLI-- 163
Query 64 SEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCWH-- 121
++E + ++LPSG + + N +G V G K L +AG+ CW
Sbjct 164 AKEGKSATLKLPSGEVRLISKNCSATVGQVGNVGVNQKSLGRAGS---------KCWLGK 214
Query 122 --KVRGVAMNPVEHPHGGG 138
VRGV MNPV+HPHGGG
Sbjct 215 RPVVRGVVMNPVDHPHGGG 233
> ath:ArthCp064 rpl2; 50S ribosomal protein L2; K02886 large subunit
ribosomal protein L2
Length=274
Score = 71.2 bits (173), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 47/139 (33%), Positives = 71/139 (51%), Gaps = 15/139 (10%)
Query 4 EGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATVVGH 63
G G I G + + +GN LPL +P G+ + ++E G G LA+ +G+ A ++
Sbjct 106 RGAIIGDTIVSGTEVPIKMGNALPLTDMPLGTAIHNIEITLGRGGQLARAAGAVAKLI-- 163
Query 64 SEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCWH-- 121
++E + ++LPSG + + N +G V G K L +AG+ CW
Sbjct 164 AKEGKSATLKLPSGEVRLISKNCSATVGQVGNVGVNQKSLGRAGS---------KCWLGK 214
Query 122 --KVRGVAMNPVEHPHGGG 138
VRGV MNPV+HPHGGG
Sbjct 215 RPVVRGVVMNPVDHPHGGG 233
> ath:AT2G44065 ribosomal protein L2 family protein
Length=214
Score = 68.6 bits (166), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 43/117 (36%), Positives = 67/117 (57%), Gaps = 6/117 (5%)
Query 22 IGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATVVGHSEETGKSRVRLPSGARKT 81
IG+ +PLG + G+++ ++E PG + + +G+ A ++ + GK ++LPSG K
Sbjct 59 IGSSMPLGMMRIGTIIHNIEMNPGQGAKMVRAAGTNAKILKEPAK-GKCLIKLPSGDTKW 117
Query 82 LPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCWHKVRGVAMNPVEHPHGGG 138
+ + R IG V+ K L KAG + ++ R KVRGVAMNP +HPHGGG
Sbjct 118 INAKCRATIGTVSNPSHGTKKLYKAGQS--RWLGIR---PKVRGVAMNPCDHPHGGG 169
> tgo:TGME49_054400 50S ribosomal protein l2, putative
Length=380
Score = 68.2 bits (165), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 44/135 (32%), Positives = 68/135 (50%), Gaps = 7/135 (5%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
LA E G + K A+++ GN LPLG +P ++V +VE +PG G + + G +ATV
Sbjct 199 LATEVTRPGDRVVASKHASIAPGNCLPLGNIPVSTIVHNVELRPGAGGQIVRAGGCYATV 258
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
V ++L S + P++ +G V+ ++ KAG +Y + R
Sbjct 259 VAKDRHFVT--LKLSSTEVRRFPADCWATVGQVSNAAHAERIRGKAGVSYWMGERPRT-- 314
Query 121 HKVRGVAMNPVEHPH 135
RG AMNPV+HPH
Sbjct 315 ---RGKAMNPVDHPH 326
> tpv:TP03_0269 60S ribosomal protein L2
Length=275
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 48/136 (35%), Positives = 67/136 (49%), Gaps = 11/136 (8%)
Query 5 GIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATVVGHS 64
G+ G + A L GN LPL +P S+V +VE +PG G +A++ G + TV
Sbjct 148 GVLPGTRLLSSSNAPLLPGNSLPLRHIPVNSIVHNVEMRPGAGGQIARSGGVYCTVTEKD 207
Query 65 EETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNC-WHK- 122
E +RL S + P + IG ++ ++ L KAGT +RN W
Sbjct 208 ETFAT--LRLSSTELRKFPLDCWATIGQISNIKHNERILKKAGT-------RRNMGWRPV 258
Query 123 VRGVAMNPVEHPHGGG 138
VRG+AMNP HPHGG
Sbjct 259 VRGIAMNPNSHPHGGN 274
> sce:YEL050C RML2; Mitochondrial ribosomal protein of the large
subunit, has similarity to E. coli L2 ribosomal protein;
fat21 mutant allele causes inability to utilize oleate and may
interfere with activity of the Adr1p transcription factor
Length=393
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 41/120 (34%), Positives = 63/120 (52%), Gaps = 13/120 (10%)
Query 23 GNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATVVGHSEETGKSRVRLPSGARKTL 82
GN LP+ +P G+++ +V P G +++G++A V+ E K+ VRL SG + +
Sbjct 238 GNCLPISMIPIGTIIHNVGITPVGPGKFCRSAGTYARVLAKLPEKKKAIVRLQSGEHRYV 297
Query 83 PSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCWH----KVRGVAMNPVEHPHGGG 138
+ IG+V+ ++ L KAG R+ W VRGVAMN +HPHGGG
Sbjct 298 SLEAVATIGVVSNIDHQNRSLGKAG---------RSRWLGIRPTVRGVAMNKCDHPHGGG 348
> ath:AT4G14250 structural constituent of ribosome
Length=724
Score = 58.5 bits (140), Expect = 8e-09, Method: Composition-based stats.
Identities = 31/67 (46%), Positives = 39/67 (58%), Gaps = 12/67 (17%)
Query 72 VRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCWHKVRGVAM-NP 130
+ LP ++KT+ S R +IG +A G K ++K RN W KVRGVAM NP
Sbjct 382 INLPLDSKKTVLSGCRVMIGQIASSGLTKKLMIK-----------RNMWAKVRGVAMMNP 430
Query 131 VEHPHGG 137
VEHPHGG
Sbjct 431 VEHPHGG 437
> bbo:BBOV_V000430 ribosomal protein L2
Length=242
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 42/131 (32%), Positives = 64/131 (48%), Gaps = 11/131 (8%)
Query 9 GQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATVVGHSEETG 68
G FI ++ + G+V L G+ + ++ L ++ S+A ++ E
Sbjct 93 GDFIKYYNRSIKTEGDVDLLDNFSVGNKIYNIYINTMQSYKLCTSALSYAIIISKYENF- 151
Query 69 KSRVRLPSGARKTLP-SNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCWHKVRGVA 127
++LPSG KT+ SNS + A +I++P K TA +K K R KVRG A
Sbjct 152 -IILKLPSG--KTIRISNSD--LASYAFSDKIEEP--KYKTAGYKIKLGRR--PKVRGTA 202
Query 128 MNPVEHPHGGG 138
MN +HPHGGG
Sbjct 203 MNACDHPHGGG 213
> tpv:TP05_0003 rpl2; ribosomal protein L2
Length=246
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 36/130 (27%), Positives = 58/130 (44%), Gaps = 9/130 (6%)
Query 9 GQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATVVGHSEETG 68
G I +K + G++ L + S+E KP + + ++ + ++ ++E
Sbjct 93 GDLIKFTEKTSKHFGDINYLNNFDINDKLYSLEFKPKIKSKICLSAKCVSILL--NKENN 150
Query 69 KSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCWHKVRGVAM 128
+++LPS K L N++C I KAGT + R KVRG AM
Sbjct 151 LIKIKLPSNKIKKL--NNKCYAVYGNSEKVISNFKQKAGT--ERLFGNR---PKVRGSAM 203
Query 129 NPVEHPHGGG 138
N +HPHGGG
Sbjct 204 NACDHPHGGG 213
> cel:F56B3.8 hypothetical protein
Length=321
Score = 37.4 bits (85), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 34/133 (25%), Positives = 58/133 (43%), Gaps = 11/133 (8%)
Query 1 LAVEGIHTGQFI----HCGKKAALSI-GNVLPLGKVPEGSVVCSVEEKPG-DRGALAKTS 54
LA E + G I H + + + GN P+G + G+V+ S+E P D K +
Sbjct 133 LATENMKAGDVISTSGHLSENPVIGVEGNAYPIGSLAAGTVINSIERYPTMDSETFVKAA 192
Query 55 GSFATVVGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHK-- 112
G+ AT+V H + + V+LP +L +G ++ ID + + + +
Sbjct 193 GTSATIVRHQGDF--TVVKLPHKHEFSLHRTCMATVGRLSHAD-IDGKIFGSAQMHRRFG 249
Query 113 YKAKRNCWHKVRG 125
YK +HK G
Sbjct 250 YKMSSGLFHKKDG 262
> hsa:51069 MRPL2, MRP-L14, RPML14; mitochondrial ribosomal protein
L2
Length=305
Score = 32.7 bits (73), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 34/135 (25%), Positives = 63/135 (46%), Gaps = 12/135 (8%)
Query 1 LAVEGIHTGQFI----HCGKKA-ALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSG 55
+A E + G I H G+ A A G+ PLG +P G+++ +VE +PG + +G
Sbjct 152 IATENMQAGDTILNSNHIGRMAVAAREGDAHPLGALPVGTLINNVESEPGRGAQYIRAAG 211
Query 56 SFATVVGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKA 115
+ ++ + G + ++LPS + + +G V+ + + KAG +++
Sbjct 212 TCGVLL--RKVNGTAIIQLPSKRQMQVLETCVATVGRVSNVDHNKRVIGKAGR--NRWLG 267
Query 116 KR---NCWHKVRGVA 127
KR WH+ G A
Sbjct 268 KRPNSGRWHRKGGWA 282
> mmu:27398 Mrpl2, CGI-22, MRP-L14, Rpml14; mitochondrial ribosomal
protein L2
Length=306
Score = 32.7 bits (73), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 31/125 (24%), Positives = 57/125 (45%), Gaps = 16/125 (12%)
Query 1 LAVEGIHTGQFI----HCGKKA-ALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSG 55
+A E + G I H G+ A A G+ PLG +P G+++ +VE +PG + +G
Sbjct 153 IATENMKAGDTILNSNHIGRMAVAAQEGDAHPLGALPVGTLINNVESEPGRGAQYIRAAG 212
Query 56 SFATVVGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKA 115
+ ++ + G + ++LPS + + + +G V+ + + KAG
Sbjct 213 TCGVLL--RKVNGTAIIQLPSKRQMQVLESCTATVGRVSNVNHNQRVIGKAG-------- 262
Query 116 KRNCW 120
RN W
Sbjct 263 -RNRW 266
> xla:447593 MGC84466 protein
Length=294
Score = 31.6 bits (70), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 21/81 (25%), Positives = 42/81 (51%), Gaps = 7/81 (8%)
Query 1 LAVEGIHTGQFI----HCGKKA-ALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSG 55
+A E + G + H G+ A + G+ PLG +P G+++ ++E +PG + +G
Sbjct 140 IATENMQEGDLLTSSGHIGRMAVSAKEGDAHPLGALPVGTLISNLEFQPGKGAQYIRAAG 199
Query 56 SFATVVGHSEETGKSRVRLPS 76
+ ++ + G + V+LPS
Sbjct 200 TSGVLL--RKVNGTAIVQLPS 218
> tpv:TP02_0314 hypothetical protein
Length=710
Score = 29.6 bits (65), Expect = 4.0, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 36 VVCSVEEKPGDRGALAKTSGSFATVVGHSEETGKSRVRLP 75
+ +++ KP D L K+SGS + G S K+RV P
Sbjct 627 ITTNLDPKPTDSSVLTKSSGSDVSKTGDSNVLAKARVTPP 666
> mmu:11519 Add2, 2900072M03Rik; adducin 2 (beta)
Length=725
Score = 28.9 bits (63), Expect = 6.3, Method: Composition-based stats.
Identities = 18/44 (40%), Positives = 21/44 (47%), Gaps = 0/44 (0%)
Query 16 KKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFAT 59
KK +S N P PEG VV EE+P AL+K G T
Sbjct 627 KKTEVSEANTEPEPVKPEGLVVNGKEEEPSVEEALSKGLGQMTT 670
> dre:393209 MGC55995; zgc:55995; K10401 kinesin family member
18/19
Length=895
Score = 28.9 bits (63), Expect = 7.3, Method: Composition-based stats.
Identities = 28/98 (28%), Positives = 42/98 (42%), Gaps = 5/98 (5%)
Query 25 VLPLGKVPEGSVVCSVEEKPG---DRGALAKTSGSFATVVGHSEETGKSRVRLPSGARKT 81
+ PL PEG S P D G + +G+ AT++ E+ G+S ++
Sbjct 718 MFPLQFTPEGQPHTSSIFDPNSTIDIGHIEDPTGANATIILSPEDPGRSTLQALRANNAN 777
Query 82 LPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNC 119
P+N I L+ R KP A T+ + K K NC
Sbjct 778 KPANRLEIPSLL--DLRRSKPSYMAMTSAAQGKRKLNC 813
Lambda K H
0.316 0.135 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3256415000
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40