bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
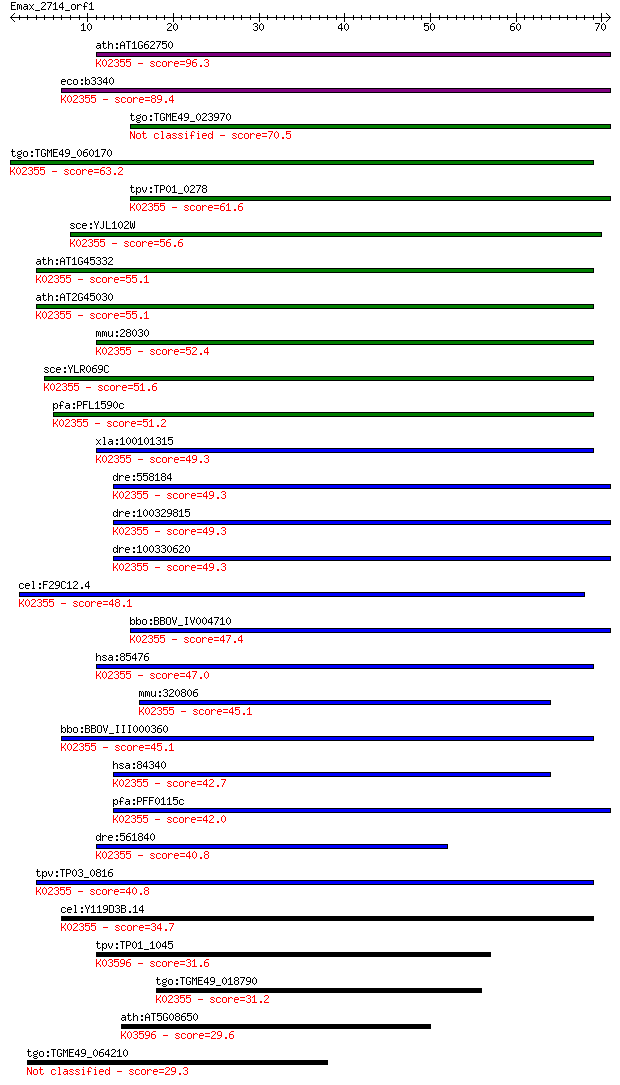
Query= Emax_2714_orf1
Length=70
Score E
Sequences producing significant alignments: (Bits) Value
ath:AT1G62750 SCO1; SCO1 (SNOWY COTYLEDON 1); ATP binding / tr... 96.3 2e-20
eco:b3340 fusA, ECK3327, far, fus, JW3302; protein chain elong... 89.4 3e-18
tgo:TGME49_023970 elongation factor G, putative (EC:3.1.6.1) 70.5 1e-12
tgo:TGME49_060170 elongation factor G, putative (EC:2.7.7.4); ... 63.2 2e-10
tpv:TP01_0278 translation elongation factor G 2; K02355 elonga... 61.6 6e-10
sce:YJL102W MEF2; Mef2p; K02355 elongation factor G 56.6 2e-08
ath:AT1G45332 mitochondrial elongation factor, putative; K0235... 55.1 6e-08
ath:AT2G45030 mitochondrial elongation factor, putative; K0235... 55.1 6e-08
mmu:28030 Gfm1, AW545374, D3Wsu133e, Gfm; G elongation factor,... 52.4 4e-07
sce:YLR069C MEF1; Mef1p; K02355 elongation factor G 51.6 6e-07
pfa:PFL1590c elongation factor G, putative; K02355 elongation ... 51.2 8e-07
xla:100101315 gfm1, EF-Gmt, coxpd1, efg, efg1, efgm, egf1, gfm... 49.3 3e-06
dre:558184 gfm2, si:dkey-35i22.3, zgc:153835; G elongation fac... 49.3 3e-06
dre:100329815 Ribosome-releasing factor 2, mitochondrial-like;... 49.3 3e-06
dre:100330620 Ribosome-releasing factor 2, mitochondrial-like;... 49.3 3e-06
cel:F29C12.4 hypothetical protein; K02355 elongation factor G 48.1 7e-06
bbo:BBOV_IV004710 23.m06266; translation elongation factor G (... 47.4 1e-05
hsa:85476 GFM1, COXPD1, EFG, EFG1, EFGM, EGF1, FLJ12662, FLJ13... 47.0 2e-05
mmu:320806 Gfm2, 6530419G12Rik, A930009M04Rik, EFG2, MST027; G... 45.1 6e-05
bbo:BBOV_III000360 17.m07056; translation elongation factor G;... 45.1 6e-05
hsa:84340 GFM2, EF-G2mt, EFG2, MRRF2, MST027, RRF2, RRF2mt, hE... 42.7 3e-04
pfa:PFF0115c elongation factor G, putative; K02355 elongation ... 42.0 5e-04
dre:561840 gfm1, zgc:154041; G elongation factor, mitochondria... 40.8 0.001
tpv:TP03_0816 elongation factor G; K02355 elongation factor G 40.8 0.001
cel:Y119D3B.14 hypothetical protein; K02355 elongation factor G 34.7 0.068
tpv:TP01_1045 hypothetical protein; K03596 GTP-binding protein... 31.6 0.67
tgo:TGME49_018790 elongation factor Tu GTP-binding domain-cont... 31.2 0.95
ath:AT5G08650 GTP-binding protein LepA, putative; K03596 GTP-b... 29.6 2.1
tgo:TGME49_064210 hypothetical protein 29.3 3.1
> ath:AT1G62750 SCO1; SCO1 (SNOWY COTYLEDON 1); ATP binding /
translation elongation factor/ translation factor, nucleic acid
binding; K02355 elongation factor G
Length=783
Score = 96.3 bits (238), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 41/60 (68%), Positives = 51/60 (85%), Gaps = 0/60 (0%)
Query 11 DDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQMHANSR 70
DD +P A LAFKI +DPFVG LTF+RVY+G + +GSYVLNA+K +ER+GRLL+MHANSR
Sbjct 391 DDDEPFAGLAFKIMSDPFVGSLTFVRVYSGKISAGSYVLNANKGKKERIGRLLEMHANSR 450
> eco:b3340 fusA, ECK3327, far, fus, JW3302; protein chain elongation
factor EF-G, GTP-binding; K02355 elongation factor G
Length=704
Score = 89.4 bits (220), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 41/64 (64%), Positives = 49/64 (76%), Gaps = 0/64 (0%)
Query 7 ELMADDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQMH 66
E A D +P +ALAFKIATDPFVG LTF RVY+G + SG VLN+ K +RER GR++QMH
Sbjct 308 ERHASDDEPFSALAFKIATDPFVGNLTFFRVYSGVVNSGDTVLNSVKAARERFGRIVQMH 367
Query 67 ANSR 70
AN R
Sbjct 368 ANKR 371
> tgo:TGME49_023970 elongation factor G, putative (EC:3.1.6.1)
Length=750
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 31/56 (55%), Positives = 39/56 (69%), Gaps = 0/56 (0%)
Query 15 PLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQMHANSR 70
PL+AL FK+ TDPFVG F+RVYTG L GS V+N ER+ RL+ +HAN+R
Sbjct 335 PLSALVFKMTTDPFVGVQNFVRVYTGELRPGSVVMNVRTGKEERIQRLVLIHANAR 390
> tgo:TGME49_060170 elongation factor G, putative (EC:2.7.7.4);
K02355 elongation factor G
Length=877
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 35/69 (50%), Positives = 45/69 (65%), Gaps = 2/69 (2%)
Query 1 KTGDEVELMADDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRER-V 59
K +EV L AD KK L ALAFKI P VG+LT++R+Y G L+ G V+N S R +
Sbjct 464 KQEEEVPLHADPKKSLVALAFKIQELP-VGQLTYLRLYQGKLKKGDSVVNVSTQKRSSPI 522
Query 60 GRLLQMHAN 68
R+LQMHA+
Sbjct 523 KRILQMHAD 531
> tpv:TP01_0278 translation elongation factor G 2; K02355 elongation
factor G
Length=803
Score = 61.6 bits (148), Expect = 6e-10, Method: Composition-based stats.
Identities = 28/56 (50%), Positives = 38/56 (67%), Gaps = 0/56 (0%)
Query 15 PLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQMHANSR 70
PLAAL FK++ D VG TFIR+Y GS+++G YV N +RV ++L MH+N R
Sbjct 418 PLAALVFKLSFDQQVGNQTFIRIYRGSIKTGDYVYNPRTKKSQRVQKILFMHSNER 473
> sce:YJL102W MEF2; Mef2p; K02355 elongation factor G
Length=819
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 26/62 (41%), Positives = 42/62 (67%), Gaps = 0/62 (0%)
Query 8 LMADDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQMHA 67
L+ ++K ALAFK+ TDP G+ FIR+Y+G+L SG+ V N++ + ++G+LL HA
Sbjct 350 LVNNNKNLCIALAFKVITDPIRGKQIFIRIYSGTLNSGNTVYNSTTGEKFKLGKLLIPHA 409
Query 68 NS 69
+
Sbjct 410 GT 411
> ath:AT1G45332 mitochondrial elongation factor, putative; K02355
elongation factor G
Length=754
Score = 55.1 bits (131), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 26/65 (40%), Positives = 41/65 (63%), Gaps = 1/65 (1%)
Query 4 DEVELMADDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLL 63
+ V L PL ALAFK+ F G+LT++RVY G ++ G +++N + R +V RL+
Sbjct 354 ERVTLTGSPDGPLVALAFKLEEGRF-GQLTYLRVYEGVIKKGDFIINVNTGKRIKVPRLV 412
Query 64 QMHAN 68
+MH+N
Sbjct 413 RMHSN 417
> ath:AT2G45030 mitochondrial elongation factor, putative; K02355
elongation factor G
Length=754
Score = 55.1 bits (131), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 26/65 (40%), Positives = 41/65 (63%), Gaps = 1/65 (1%)
Query 4 DEVELMADDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLL 63
+ V L PL ALAFK+ F G+LT++RVY G ++ G +++N + R +V RL+
Sbjct 354 ERVTLTGSPDGPLVALAFKLEEGRF-GQLTYLRVYEGVIKKGDFIINVNTGKRIKVPRLV 412
Query 64 QMHAN 68
+MH+N
Sbjct 413 RMHSN 417
> mmu:28030 Gfm1, AW545374, D3Wsu133e, Gfm; G elongation factor,
mitochondrial 1; K02355 elongation factor G
Length=751
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 25/58 (43%), Positives = 35/58 (60%), Gaps = 1/58 (1%)
Query 11 DDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQMHAN 68
DD P LAFK+ F G+LT++R Y G L+ GS + N + RV RL++MHA+
Sbjct 348 DDSHPFVGLAFKLEAGRF-GQLTYVRNYQGELKKGSTIYNTRTGKKVRVQRLVRMHAD 404
> sce:YLR069C MEF1; Mef1p; K02355 elongation factor G
Length=761
Score = 51.6 bits (122), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 40/64 (62%), Gaps = 1/64 (1%)
Query 5 EVELMADDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQ 64
+V L+ ++P LAFK+ + G+LT++RVY G L G+Y+ N + +V RL++
Sbjct 364 KVNLVPAVQQPFVGLAFKLEEGKY-GQLTYVRVYQGRLRKGNYITNVKTGKKVKVARLVR 422
Query 65 MHAN 68
MH++
Sbjct 423 MHSS 426
> pfa:PFL1590c elongation factor G, putative; K02355 elongation
factor G
Length=803
Score = 51.2 bits (121), Expect = 8e-07, Method: Composition-based stats.
Identities = 18/63 (28%), Positives = 37/63 (58%), Gaps = 0/63 (0%)
Query 6 VELMADDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQM 65
++L+ D+ P+ FKI D G++++ R+Y G ++ + N N +E V ++++M
Sbjct 405 IQLLCDNNLPMVGFLFKIQEDNMYGQMSYFRIYQGKIKKKEMITNMMTNKKEIVKKIMKM 464
Query 66 HAN 68
H+N
Sbjct 465 HSN 467
> xla:100101315 gfm1, EF-Gmt, coxpd1, efg, efg1, efgm, egf1, gfm;
G elongation factor, mitochondrial 1; K02355 elongation
factor G
Length=748
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 23/58 (39%), Positives = 33/58 (56%), Gaps = 1/58 (1%)
Query 11 DDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQMHAN 68
D +P LAFK+ F G+LT++RVY G L Y+ N + RV RL+ +HA+
Sbjct 345 DSSQPFVGLAFKLEAGRF-GQLTYVRVYQGMLRKSDYIYNTRTGKKVRVQRLVCLHAD 401
> dre:558184 gfm2, si:dkey-35i22.3, zgc:153835; G elongation factor,
mitochondrial 2; K02355 elongation factor G
Length=762
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 24/58 (41%), Positives = 37/58 (63%), Gaps = 0/58 (0%)
Query 13 KKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQMHANSR 70
K L ALAFK+ D G L F+R+Y+GS+++ S V N ++N E++ RLL A+ +
Sbjct 352 KNDLCALAFKVVHDKQRGPLVFVRIYSGSMKAQSSVHNINRNETEKMSRLLLPFADQQ 409
> dre:100329815 Ribosome-releasing factor 2, mitochondrial-like;
K02355 elongation factor G
Length=762
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 24/58 (41%), Positives = 37/58 (63%), Gaps = 0/58 (0%)
Query 13 KKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQMHANSR 70
K L ALAFK+ D G L F+R+Y+GS+++ S V N ++N E++ RLL A+ +
Sbjct 352 KNDLCALAFKVVHDKQRGPLVFVRIYSGSMKAQSSVHNINRNETEKMSRLLLPFADQQ 409
> dre:100330620 Ribosome-releasing factor 2, mitochondrial-like;
K02355 elongation factor G
Length=684
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 24/58 (41%), Positives = 37/58 (63%), Gaps = 0/58 (0%)
Query 13 KKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQMHANSR 70
K L ALAFK+ D G L F+R+Y+GS+++ S V N ++N E++ RLL A+ +
Sbjct 352 KNDLCALAFKVVHDKQRGPLVFVRIYSGSMKAQSSVHNINRNETEKMSRLLLPFADQQ 409
> cel:F29C12.4 hypothetical protein; K02355 elongation factor
G
Length=750
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 28/70 (40%), Positives = 38/70 (54%), Gaps = 5/70 (7%)
Query 2 TGDEVELMADDK----KPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRE 57
TGDE ++ K KP LAFK+ + G+LT+ RVY G L G V + +
Sbjct 332 TGDEKGIILSPKRNNDKPFVGLAFKLEAGKY-GQLTYFRVYQGQLSKGDTVYASRDGRKV 390
Query 58 RVGRLLQMHA 67
RV RL++MHA
Sbjct 391 RVQRLVRMHA 400
> bbo:BBOV_IV004710 23.m06266; translation elongation factor G
(EF-G); K02355 elongation factor G
Length=817
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 21/56 (37%), Positives = 31/56 (55%), Gaps = 0/56 (0%)
Query 15 PLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQMHANSR 70
PLA L FKI+ D +G ++R+Y G + G V N +V +LL +H+N R
Sbjct 429 PLAGLVFKISHDAQIGTQAYVRIYRGQVSVGDVVYNPRTKKSNKVQKLLFIHSNER 484
> hsa:85476 GFM1, COXPD1, EFG, EFG1, EFGM, EGF1, FLJ12662, FLJ13632,
FLJ20773, GFM, hEFG1; G elongation factor, mitochondrial
1; K02355 elongation factor G
Length=751
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 22/58 (37%), Positives = 33/58 (56%), Gaps = 1/58 (1%)
Query 11 DDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQMHAN 68
D+ P LAFK+ F G+LT++R Y G L+ G + N + R+ RL +MHA+
Sbjct 348 DNSHPFVGLAFKLEVGRF-GQLTYVRSYQGELKKGDTIYNTRTRKKVRLQRLARMHAD 404
> mmu:320806 Gfm2, 6530419G12Rik, A930009M04Rik, EFG2, MST027;
G elongation factor, mitochondrial 2; K02355 elongation factor
G
Length=741
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 22/48 (45%), Positives = 30/48 (62%), Gaps = 0/48 (0%)
Query 16 LAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLL 63
L ALAFK+ D G L F+R+Y+G+L V N ++N ER+ RLL
Sbjct 367 LCALAFKVLHDKQRGPLVFLRIYSGTLTPQLAVHNINRNCTERMSRLL 414
> bbo:BBOV_III000360 17.m07056; translation elongation factor
G; K02355 elongation factor G
Length=741
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 34/62 (54%), Gaps = 1/62 (1%)
Query 7 ELMADDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQMH 66
EL K+PL A AFKI P +G+LTF+R+Y G + G + + +L +MH
Sbjct 343 ELDGGYKQPLVAYAFKIQDSP-MGQLTFLRLYQGMMRRGQQLYLVEDGKKHSTKKLFKMH 401
Query 67 AN 68
A+
Sbjct 402 AS 403
> hsa:84340 GFM2, EF-G2mt, EFG2, MRRF2, MST027, RRF2, RRF2mt,
hEFG2, mEF-G_2; G elongation factor, mitochondrial 2; K02355
elongation factor G
Length=779
Score = 42.7 bits (99), Expect = 3e-04, Method: Composition-based stats.
Identities = 21/51 (41%), Positives = 31/51 (60%), Gaps = 0/51 (0%)
Query 13 KKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLL 63
K L ALAFK+ D G L F+R+Y+G+++ + N + N ER+ RLL
Sbjct 364 KDDLCALAFKVLHDKQRGPLVFMRIYSGTIKPQLAIHNINGNCTERISRLL 414
> pfa:PFF0115c elongation factor G, putative; K02355 elongation
factor G
Length=937
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 13/58 (22%), Positives = 32/58 (55%), Gaps = 0/58 (0%)
Query 13 KKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQMHANSR 70
K+ + L +KI D +G + ++R+Y G + G ++ N E++ ++ +H++ +
Sbjct 505 KRKMVGLIYKIMNDQHLGNINYVRIYEGQINRGEFIYNNRTKKSEKISKIFFIHSSEK 562
> dre:561840 gfm1, zgc:154041; G elongation factor, mitochondrial
1; K02355 elongation factor G
Length=745
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 18/41 (43%), Positives = 25/41 (60%), Gaps = 1/41 (2%)
Query 11 DDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNA 51
DD +P LAFK+ F G+LT++RVY G L Y+ N+
Sbjct 342 DDTQPFVGLAFKLEAGRF-GQLTYVRVYQGCLRKTDYIHNS 381
> tpv:TP03_0816 elongation factor G; K02355 elongation factor
G
Length=805
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 22/65 (33%), Positives = 40/65 (61%), Gaps = 2/65 (3%)
Query 4 DEVELMADDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLL 63
+++EL +DK L FKI D ++G+L++IR+Y G L G VL ++ R + +L
Sbjct 405 NKIELKPEDKG-LVGYIFKI-VDTYLGQLSYIRIYKGVLRRGLSVLVVEEDKRVTLKKLY 462
Query 64 QMHAN 68
++H++
Sbjct 463 KVHSD 467
> cel:Y119D3B.14 hypothetical protein; K02355 elongation factor
G
Length=689
Score = 34.7 bits (78), Expect = 0.068, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 32/62 (51%), Gaps = 0/62 (0%)
Query 7 ELMADDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQMH 66
EL K + I D G+L+++R+YTGSL + S + N S+ + E +L +
Sbjct 264 ELTCASKIATISCGSAITHDKRRGQLSYMRIYTGSLHNNSTIFNTSQMTSEGPLKLFTPY 323
Query 67 AN 68
A+
Sbjct 324 AD 325
> tpv:TP01_1045 hypothetical protein; K03596 GTP-binding protein
LepA
Length=711
Score = 31.6 bits (70), Expect = 0.67, Method: Composition-based stats.
Identities = 17/48 (35%), Positives = 27/48 (56%), Gaps = 2/48 (4%)
Query 11 DDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYV--LNASKNSR 56
D +KP AL F D + G + +IRV+ GS + G + +NA+ S+
Sbjct 279 DLEKPFRALVFDSFYDSYKGAICYIRVFEGSAKVGDEITLMNANITSK 326
> tgo:TGME49_018790 elongation factor Tu GTP-binding domain-containing
protein (EC:2.7.7.4); K02355 elongation factor G
Length=1133
Score = 31.2 bits (69), Expect = 0.95, Method: Compositional matrix adjust.
Identities = 13/38 (34%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 18 ALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNS 55
+AFK+ D GR+ F+RV G + + + N++KN
Sbjct 548 CVAFKVYPDGKGGRIAFVRVLAGRITPRTSLFNSTKNC 585
> ath:AT5G08650 GTP-binding protein LepA, putative; K03596 GTP-binding
protein LepA
Length=681
Score = 29.6 bits (65), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 19/36 (52%), Gaps = 0/36 (0%)
Query 14 KPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVL 49
KPL AL F DP+ G + + RV G ++ G +
Sbjct 270 KPLRALIFDSYYDPYRGVIVYFRVIDGKVKKGDRIF 305
> tgo:TGME49_064210 hypothetical protein
Length=1185
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 3 GDEVELMADDKKPLAALAFKIATDPFVGRLTFIRV 37
G+E L +KPL+ LA K + DP R+T R
Sbjct 133 GEEKTLSIRKRKPLSELALKDSEDPAAARVTPARC 167
Lambda K H
0.317 0.132 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2024947620
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40