bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2675_orf1
Length=142
Score E
Sequences producing significant alignments: (Bits) Value
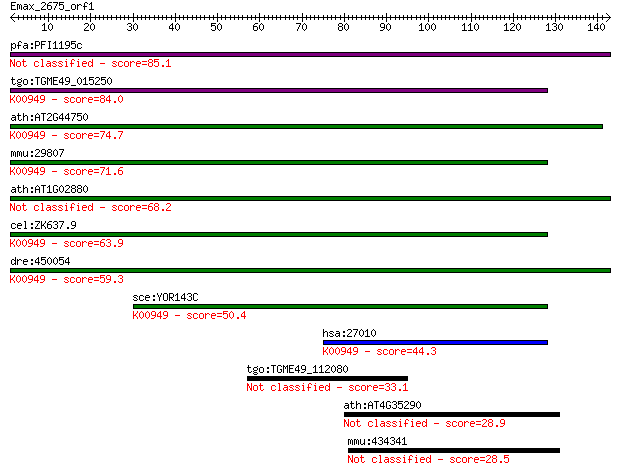
pfa:PFI1195c thiamin pyrophosphokinase, putative (EC:2.7.6.2) 85.1 6e-17
tgo:TGME49_015250 thiamin pyrophosphokinase, putative (EC:2.7.... 84.0 1e-16
ath:AT2G44750 TPK2; TPK2 (Thiamin pyrophosphokinase 2); thiami... 74.7 9e-14
mmu:29807 Tpk1; thiamine pyrophosphokinase (EC:2.7.6.2); K0094... 71.6 8e-13
ath:AT1G02880 TPK1; TPK1 (THIAMIN PYROPHOSPHOKINASE1); thiamin... 68.2 9e-12
cel:ZK637.9 tpk-1; Thiamine PyrophosphoKinase family member (t... 63.9 2e-10
dre:450054 tpk1, zgc:101685; thiamin pyrophosphokinase 1 (EC:2... 59.3 4e-09
sce:YOR143C THI80; Thi80p (EC:2.7.6.2); K00949 thiamine pyroph... 50.4 2e-06
hsa:27010 TPK1, HTPK1, PP20; thiamin pyrophosphokinase 1 (EC:2... 44.3 1e-04
tgo:TGME49_112080 hypothetical protein 33.1 0.27
ath:AT4G35290 GLUR2; GLUR2 (GLUTAMATE RECEPTOR 2); intracellul... 28.9 6.0
mmu:434341 Nlrc5, AI451557, AK220210, NOD27; NLR family, CARD ... 28.5 6.9
> pfa:PFI1195c thiamin pyrophosphokinase, putative (EC:2.7.6.2)
Length=400
Score = 85.1 bits (209), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 52/142 (36%), Positives = 81/142 (57%), Gaps = 8/142 (5%)
Query 1 DQIHTDLEKAFAFAASKFRLSHDDIVIIAGAIGGRFDHSITAVSFLYKVHNASAETLPPK 60
+Q +TDL+K K + +D +++ GA G RFD + +S LYK + +T+
Sbjct 256 NQENTDLDKCIE--KIKPYIYENDKILVLGATGNRFDQTCANISSLYK----NVQTIN-N 308
Query 61 VILLGECNACLLLPEGDNEVLLSDGVFSEACGLLPMAGEVRNVTTEGLKWNVKGEPLRVG 120
+ L+GE N LL +G++ + ++ F + C LLP+ G+ + V TEGLK+N+ E L
Sbjct 309 IYLIGENNFIFLLKKGNHVIQINLNAFQKGCALLPIGGKCK-VKTEGLKYNLNYEYLSFD 367
Query 121 GLISSSNIRIKVRIKIATSDPL 142
LISSSN I+ IKI+ PL
Sbjct 368 SLISSSNEIIQNEIKISNDTPL 389
> tgo:TGME49_015250 thiamin pyrophosphokinase, putative (EC:2.7.6.2);
K00949 thiamine pyrophosphokinase [EC:2.7.6.2]
Length=460
Score = 84.0 bits (206), Expect = 1e-16, Method: Composition-based stats.
Identities = 55/159 (34%), Positives = 80/159 (50%), Gaps = 32/159 (20%)
Query 1 DQIHTDLEKAFAFAASKFRLSHDDIVIIAGAIGGRFDHSITAVSFLYKVH---------- 50
DQ D+EKA+ + R S +D+VII GAIGGR DH++ A+ LYK+
Sbjct 250 DQDLPDVEKAWRLLLAPKRFSSNDVVIILGAIGGRLDHTLCAIHVLYKLTAEHEAAEARA 309
Query 51 -------------------NAS--AETLPPKVILLGECNACLLLPEGDNEVLLSDGVFSE 89
NA+ ++ L ++ LLGE + C L+ +G V+ SD + +
Sbjct 310 KAVEKREGNGEESREGQRGNATPASDFLCFQIYLLGEDSLCFLVSKGRTRVIPSDLLITR 369
Query 90 ACGLLPMAGEVRNVTTEGLKWNVKGE-PLRVGGLISSSN 127
C L+P V VTTEGL+WN+ + L G IS+SN
Sbjct 370 QCALIPCGEAVSGVTTEGLRWNLTPDMRLNFGEFISTSN 408
> ath:AT2G44750 TPK2; TPK2 (Thiamin pyrophosphokinase 2); thiamin
diphosphokinase (EC:2.7.6.2); K00949 thiamine pyrophosphokinase
[EC:2.7.6.2]
Length=265
Score = 74.7 bits (182), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 45/141 (31%), Positives = 72/141 (51%), Gaps = 9/141 (6%)
Query 1 DQIHTDLEKAFAFAASKFRLSHDDIVIIAGAIGGRFDHSITAVSFLYKVHNASAETLPPK 60
DQ TDL+K ++ ++ GA+GGRFDH ++ LY+ + +
Sbjct 112 DQDTTDLDKCISYIRHSTLNQESSRILATGALGGRFDHEAGNLNVLYRYPDT-------R 164
Query 61 VILLGECNACLLLPEGD-NEVLLSDGVFSEACGLLPMAGEVRNVTTEGLKWNVKGEPLRV 119
++LL + LLP+ +E+ + + CGL+P+ N TT GLKW++ +R
Sbjct 165 IVLLSDDCLIQLLPKTHRHEIHIHSSLQGPHCGLIPIGTPSANTTTSGLKWDLSNTEMRF 224
Query 120 GGLISSSNIRIKVRIKIATSD 140
GGLIS+SN+ +K I SD
Sbjct 225 GGLISTSNL-VKEEIITVESD 244
> mmu:29807 Tpk1; thiamine pyrophosphokinase (EC:2.7.6.2); K00949
thiamine pyrophosphokinase [EC:2.7.6.2]
Length=243
Score = 71.6 bits (174), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 40/129 (31%), Positives = 66/129 (51%), Gaps = 6/129 (4%)
Query 1 DQIHTDLEKAFAFAASKFRLS--HDDIVIIAGAIGGRFDHSITAVSFLYKVHNASAETLP 58
DQ HTD K K D+++ G +GGRFD + +V+ L++ + + P
Sbjct 95 DQDHTDFTKCLQVLQRKIEEKELQVDVIVTLGGLGGRFDQIMASVNTLFQATHIT----P 150
Query 59 PKVILLGECNACLLLPEGDNEVLLSDGVFSEACGLLPMAGEVRNVTTEGLKWNVKGEPLR 118
+I++ + + LL G + + + G+ CGL+P+ VTT GLKWN+ + L
Sbjct 151 VPIIIIQKDSLIYLLQPGKHRLHVDTGMEGSWCGLIPVGQPCNQVTTTGLKWNLTNDVLG 210
Query 119 VGGLISSSN 127
G L+S+SN
Sbjct 211 FGTLVSTSN 219
> ath:AT1G02880 TPK1; TPK1 (THIAMIN PYROPHOSPHOKINASE1); thiamin
diphosphokinase
Length=180
Score = 68.2 bits (165), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 44/145 (30%), Positives = 74/145 (51%), Gaps = 10/145 (6%)
Query 1 DQIHTDLEKAFAF-AASKFRLSHDDIVIIA-GAIGGRFDHSITAVSFLYKVHNASAETLP 58
DQ TDL+K + S + I+A GA+GGRFDH ++ LY+ +
Sbjct 25 DQDTTDLDKCILYIRHSTLNQETSGLQILATGALGGRFDHEAGNLNVLYRYPDT------ 78
Query 59 PKVILLGECNACLLLPEGD-NEVLLSDGVFSEACGLLPMAGEVRNVTTEGLKWNVKGEPL 117
+++LL + LLP+ +E+ + + CGL+P+ TT GL+W++ +
Sbjct 79 -RIVLLSDDCLIQLLPKTHRHEIHIQSSLEGPHCGLIPIGTPSAKTTTSGLQWDLSNTEM 137
Query 118 RVGGLISSSNIRIKVRIKIATSDPL 142
R GGLIS+SN+ + +I + + L
Sbjct 138 RFGGLISTSNLVKEEKITVESDSDL 162
> cel:ZK637.9 tpk-1; Thiamine PyrophosphoKinase family member
(tpk-1); K00949 thiamine pyrophosphokinase [EC:2.7.6.2]
Length=243
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 37/129 (28%), Positives = 73/129 (56%), Gaps = 7/129 (5%)
Query 1 DQIHTDLEKAFAFAASKFRLSHDDI--VIIAGAIGGRFDHSITAVSFLYKVHNASAETLP 58
DQ +TDL K+ + + L+ + +++ G + GRFDH+++ +S L + ++
Sbjct 89 DQDYTDLSKSVQWCLEQKTLTSWEFENIVVLGGLNGRFDHTMSTLSSLIRFVDSQT---- 144
Query 59 PKVILLGECNACLLLPEGDNEVLLSDGVFSEACGLLPMAGEVRNVTTEGLKWNVKGEPLR 118
VI+L N L +P GD+ + ++ + ++ CG++P+ + V++ GLK+ ++ L
Sbjct 145 -PVIVLDSRNLVLAVPTGDSNLDVNLEMTTKMCGIIPIVQKETIVSSIGLKYEMENLALE 203
Query 119 VGGLISSSN 127
G LIS+SN
Sbjct 204 FGKLISTSN 212
> dre:450054 tpk1, zgc:101685; thiamin pyrophosphokinase 1 (EC:2.7.6.2);
K00949 thiamine pyrophosphokinase [EC:2.7.6.2]
Length=257
Score = 59.3 bits (142), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 48/153 (31%), Positives = 74/153 (48%), Gaps = 17/153 (11%)
Query 1 DQIHTDLEKAFAFAASKFR---LSHDDIVIIAGAIGGRFDHSITAVSFLYKVHNASAETL 57
DQ TD K A + + L D IV + G +GGRFD ++ L+ H L
Sbjct 95 DQDLTDFTKCLAIMLEEIKAKKLQIDSIVTLGG-LGGRFDQTMATEETLF--HAQKMTDL 151
Query 58 PPKVILLGECNACLLLPEGDNEVL-LSDGVFSEACGLLPMAGEVRNVTTEGLKWNVKGEP 116
P V+++ + + LL EG + L ++ G+ + C L+P+ G TT GLKWN+ +
Sbjct 152 P--VVVIQDSSLAFLLKEGRHHQLNVNTGMEGKWCSLVPV-GSPCLTTTSGLKWNLDNQV 208
Query 117 LRVGGLISSSNIR-------IKVRIKIATSDPL 142
L G L+S+SN + + I T +PL
Sbjct 209 LAFGQLVSTSNTYEDHDPKDCRKPVTITTDNPL 241
> sce:YOR143C THI80; Thi80p (EC:2.7.6.2); K00949 thiamine pyrophosphokinase
[EC:2.7.6.2]
Length=319
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 39/106 (36%), Positives = 54/106 (50%), Gaps = 15/106 (14%)
Query 30 GAIGGRFDHSITAVSFLYKV-HNASAETL----PPKVILLGECNACLLL--PEGDNEVLL 82
G IGGRFD ++ +++ LY + NAS L P +I L + N L+ P+ N +
Sbjct 190 GGIGGRFDQTVHSITQLYTLSENASYFKLCYMTPTDLIFLIKKNGTLIEYDPQFRNTCIG 249
Query 83 SDGVFSEACGLLPMAGEVRNVTTEGLKWNVKGEPLR-VGGLISSSN 127
+ CGLLP+ T GLKW+VK P V G +SSSN
Sbjct 250 N-------CGLLPIGEATLVKETRGLKWDVKNWPTSVVTGRVSSSN 288
> hsa:27010 TPK1, HTPK1, PP20; thiamin pyrophosphokinase 1 (EC:2.7.6.2);
K00949 thiamine pyrophosphokinase [EC:2.7.6.2]
Length=194
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 32/53 (60%), Gaps = 0/53 (0%)
Query 75 EGDNEVLLSDGVFSEACGLLPMAGEVRNVTTEGLKWNVKGEPLRVGGLISSSN 127
+G + + + G+ + CGL+P+ VTT GLKWN+ + L G L+S+SN
Sbjct 118 KGKHRLHVDTGMEGDWCGLIPVGQPCMQVTTTGLKWNLTNDVLAFGTLVSTSN 170
> tgo:TGME49_112080 hypothetical protein
Length=766
Score = 33.1 bits (74), Expect = 0.27, Method: Composition-based stats.
Identities = 17/40 (42%), Positives = 22/40 (55%), Gaps = 2/40 (5%)
Query 57 LPPKVI--LLGECNACLLLPEGDNEVLLSDGVFSEACGLL 94
LPP + + GE LLP GD+ LL DG+F + GL
Sbjct 549 LPPSFLSSVFGESAGDSLLPGGDSSFLLEDGLFRDDAGLF 588
> ath:AT4G35290 GLUR2; GLUR2 (GLUTAMATE RECEPTOR 2); intracellular
ligand-gated ion channel
Length=912
Score = 28.9 bits (63), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 30/64 (46%), Gaps = 17/64 (26%)
Query 80 VLLSDGVFSEACGLLP-------------MAGEVRNVTTEGLKWNVKGEPLRVGGLISSS 126
VL+ DG+ SE GL P + GEV N+ + + +V +P +GG S
Sbjct 12 VLIGDGMISEGAGLRPRYVDVGAIFSLGTLQGEVTNIAMKAAEEDVNSDPSFLGG----S 67
Query 127 NIRI 130
+RI
Sbjct 68 KLRI 71
> mmu:434341 Nlrc5, AI451557, AK220210, NOD27; NLR family, CARD
domain containing 5
Length=1915
Score = 28.5 bits (62), Expect = 6.9, Method: Composition-based stats.
Identities = 15/50 (30%), Positives = 25/50 (50%), Gaps = 0/50 (0%)
Query 81 LLSDGVFSEACGLLPMAGEVRNVTTEGLKWNVKGEPLRVGGLISSSNIRI 130
LL D V +E +LP G+++ V E + +G L GL+ S + +
Sbjct 1833 LLGDEVAAELAQVLPQMGQLKKVNLEWNRITARGAQLLAQGLVQGSCVPV 1882
Lambda K H
0.320 0.138 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2683748972
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40