bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
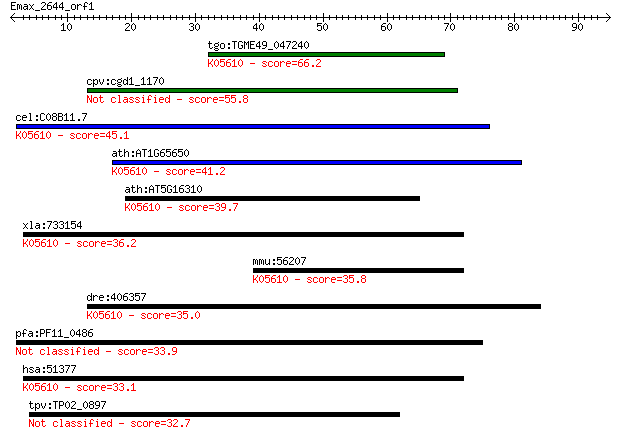
Query= Emax_2644_orf1
Length=94
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_047240 ubiquitin carboxyl-terminal hydrolase isozym... 66.2 2e-11
cpv:cgd1_1170 ubiquitin C-terminal hydrolase 55.8 4e-08
cel:C08B11.7 ubh-4; UBiquitin C-terminal Hydrolase (family 1) ... 45.1 5e-05
ath:AT1G65650 UCH2; UCH2; ubiquitin thiolesterase/ ubiquitin-s... 41.2 8e-04
ath:AT5G16310 UCH1; UCH1; ubiquitin thiolesterase; K05610 ubiq... 39.7 0.002
xla:733154 uchl5; ubiquitin carboxyl-terminal hydrolase L5 (EC... 36.2 0.025
mmu:56207 Uchl5, 5830413B11Rik, Uch37; ubiquitin carboxyl-term... 35.8 0.034
dre:406357 uchl5, wu:fj17f09, zgc:85615; ubiquitin carboxyl-te... 35.0 0.062
pfa:PF11_0486 MAEBL, putative 33.9 0.12
hsa:51377 UCHL5, CGI-70, INO80R, UCH-L5, UCH37; ubiquitin carb... 33.1 0.20
tpv:TP02_0897 hypothetical protein 32.7 0.26
> tgo:TGME49_047240 ubiquitin carboxyl-terminal hydrolase isozyme
L5, putative (EC:3.4.19.12); K05610 ubiquitin carboxyl-terminal
hydrolase L5 [EC:3.4.19.12]
Length=407
Score = 66.2 bits (160), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 27/37 (72%), Positives = 32/37 (86%), Gaps = 0/37 (0%)
Query 32 KRAEWDRENARRRHDFTPFVLCALRHLARKGELVKAV 68
KRA W +ENARRRHDF PF+L ++HLARKGELVK+V
Sbjct 345 KRARWKKENARRRHDFVPFLLTVIKHLARKGELVKSV 381
> cpv:cgd1_1170 ubiquitin C-terminal hydrolase
Length=398
Score = 55.8 bits (133), Expect = 4e-08, Method: Composition-based stats.
Identities = 28/62 (45%), Positives = 39/62 (62%), Gaps = 4/62 (6%)
Query 13 QAALSKI--EVLEELKALEQAK--RAEWDRENARRRHDFTPFVLCALRHLARKGELVKAV 68
Q ++KI E+ E L ++ K R W +EN RRRHDF PFVL LRH ++KG L+K +
Sbjct 335 QKEITKITHEISENLSIIQNEKYNREIWKKENERRRHDFLPFVLTLLRHASKKGLLMKKL 394
Query 69 RR 70
+
Sbjct 395 SQ 396
> cel:C08B11.7 ubh-4; UBiquitin C-terminal Hydrolase (family 1)
family member (ubh-4); K05610 ubiquitin carboxyl-terminal
hydrolase L5 [EC:3.4.19.12]
Length=321
Score = 45.1 bits (105), Expect = 5e-05, Method: Composition-based stats.
Identities = 29/74 (39%), Positives = 42/74 (56%), Gaps = 7/74 (9%)
Query 2 AEEAKQLEEARQAALSKIEVLEELKALEQAKRAEWDRENARRRHDFTPFVLCALRHLARK 61
A E +LEE +I L + A E K + +EN RRRH++TPFV+ ++ LA++
Sbjct 236 ANENNELEE-------QIADLNKAIADEDYKMEMYRKENNRRRHNYTPFVIELMKILAKE 288
Query 62 GELVKAVRRATAAA 75
G+LV V A AA
Sbjct 289 GKLVGLVDNAYQAA 302
> ath:AT1G65650 UCH2; UCH2; ubiquitin thiolesterase/ ubiquitin-specific
protease; K05610 ubiquitin carboxyl-terminal hydrolase
L5 [EC:3.4.19.12]
Length=330
Score = 41.2 bits (95), Expect = 8e-04, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 34/64 (53%), Gaps = 0/64 (0%)
Query 17 SKIEVLEELKALEQAKRAEWDRENARRRHDFTPFVLCALRHLARKGELVKAVRRATAAAA 76
S IE + +E+ K +W EN RR+H++ PF+ L+ LA K +L + +A
Sbjct 267 SGIEAASDKIVMEEEKFMKWRTENIRRKHNYIPFLFNFLKLLAEKKQLKPLIEKAKKQKT 326
Query 77 ATAT 80
++T
Sbjct 327 ESST 330
> ath:AT5G16310 UCH1; UCH1; ubiquitin thiolesterase; K05610 ubiquitin
carboxyl-terminal hydrolase L5 [EC:3.4.19.12]
Length=334
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 28/46 (60%), Gaps = 0/46 (0%)
Query 19 IEVLEELKALEQAKRAEWDRENARRRHDFTPFVLCALRHLARKGEL 64
IE + + +E+ K W +EN RR+H++ PF+ L+ LA K +L
Sbjct 280 IETVSQKIVMEEEKSKNWKKENMRRKHNYVPFLFNFLKILADKKKL 325
> xla:733154 uchl5; ubiquitin carboxyl-terminal hydrolase L5 (EC:3.4.19.12);
K05610 ubiquitin carboxyl-terminal hydrolase
L5 [EC:3.4.19.12]
Length=329
Score = 36.2 bits (82), Expect = 0.025, Method: Composition-based stats.
Identities = 20/69 (28%), Positives = 39/69 (56%), Gaps = 3/69 (4%)
Query 3 EEAKQLEEARQAALSKIEVLEELKALEQAKRAEWDRENARRRHDFTPFVLCALRHLARKG 62
++ L + Q+ ++K ++L E E+ K + EN RR+H++ PF++ L+ LA
Sbjct 251 DQGSTLLSSMQSEIAKYQLLIEE---EKQKMKRYKVENIRRKHNYLPFIMELLKTLAEHQ 307
Query 63 ELVKAVRRA 71
+L+ V +A
Sbjct 308 QLIPLVEKA 316
> mmu:56207 Uchl5, 5830413B11Rik, Uch37; ubiquitin carboxyl-terminal
esterase L5 (EC:3.4.19.12); K05610 ubiquitin carboxyl-terminal
hydrolase L5 [EC:3.4.19.12]
Length=328
Score = 35.8 bits (81), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 22/33 (66%), Gaps = 0/33 (0%)
Query 39 ENARRRHDFTPFVLCALRHLARKGELVKAVRRA 71
EN RR+H++ PF++ L+ LA +L+ V +A
Sbjct 283 ENIRRKHNYLPFIMELLKTLAEHQQLIPLVEKA 315
> dre:406357 uchl5, wu:fj17f09, zgc:85615; ubiquitin carboxyl-terminal
hydrolase L5 (EC:3.4.19.12); K05610 ubiquitin carboxyl-terminal
hydrolase L5 [EC:3.4.19.12]
Length=329
Score = 35.0 bits (79), Expect = 0.062, Method: Composition-based stats.
Identities = 21/71 (29%), Positives = 37/71 (52%), Gaps = 3/71 (4%)
Query 13 QAALSKIEVLEELKALEQAKRAEWDRENARRRHDFTPFVLCALRHLARKGELVKAVRRAT 72
Q+ ++K ++L E E K + EN RR+H++ PF++ L+ LA +L+ V +A
Sbjct 261 QSEIAKYQLLIEE---ENQKLKRYKVENIRRKHNYLPFIMELLKTLAEYQQLIPLVEKAK 317
Query 73 AAAAATATAAA 83
+A A
Sbjct 318 EKQSAKKIQEA 328
> pfa:PF11_0486 MAEBL, putative
Length=2055
Score = 33.9 bits (76), Expect = 0.12, Method: Composition-based stats.
Identities = 31/83 (37%), Positives = 46/83 (55%), Gaps = 12/83 (14%)
Query 2 AEEAKQLEEARQAALSKIEVLEELKALEQAKRAEWDR--ENAR-----RRHDFTPFVLCA 54
AE+A+++EEAR+A ++ +EE + E A+R E R E+AR RR + + A
Sbjct 1108 AEDARRIEEARRAEDAR--RIEEARRAEDARRVEIARRVEDARRIEISRRAEDAKRIEAA 1165
Query 55 LRHL-ARKGELVKA--VRRATAA 74
R + R+ EL KA RR AA
Sbjct 1166 RRAIEVRRAELRKAEDARRIEAA 1188
> hsa:51377 UCHL5, CGI-70, INO80R, UCH-L5, UCH37; ubiquitin carboxyl-terminal
hydrolase L5 (EC:3.4.19.12); K05610 ubiquitin
carboxyl-terminal hydrolase L5 [EC:3.4.19.12]
Length=328
Score = 33.1 bits (74), Expect = 0.20, Method: Composition-based stats.
Identities = 20/69 (28%), Positives = 42/69 (60%), Gaps = 3/69 (4%)
Query 3 EEAKQLEEARQAALSKIEVLEELKALEQAKRAEWDRENARRRHDFTPFVLCALRHLARKG 62
++ + A Q+ ++K ++L E + +++ KR + EN RR+H++ PF++ L+ LA
Sbjct 250 DQGNSMLSAIQSEVAKNQMLIE-EEVQKLKR--YKIENIRRKHNYLPFIMELLKTLAEHQ 306
Query 63 ELVKAVRRA 71
+L+ V +A
Sbjct 307 QLIPLVEKA 315
> tpv:TP02_0897 hypothetical protein
Length=1499
Score = 32.7 bits (73), Expect = 0.26, Method: Composition-based stats.
Identities = 18/58 (31%), Positives = 29/58 (50%), Gaps = 4/58 (6%)
Query 4 EAKQLEEARQAALSKIEVLEELKALEQAKRAEWDRENARRRHDFTPFVLCALRHLARK 61
E +L+ R+ AL +E + E+ KR+ +DRE+ R+ F F L LA +
Sbjct 1215 EVDRLQTLREQALKIVETFVQ----EEVKRSNYDRESIRKLKQFFDFQTLVLSTLAER 1268
Lambda K H
0.313 0.122 0.325
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2069995292
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40