bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2612_orf2
Length=99
Score E
Sequences producing significant alignments: (Bits) Value
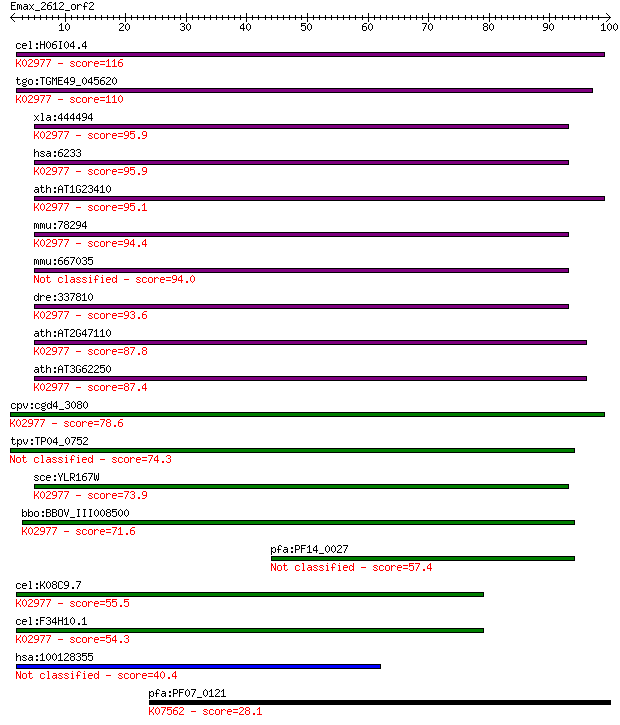
cel:H06I04.4 ubl-1; UBiquitin-Like family member (ubl-1); K029... 116 2e-26
tgo:TGME49_045620 ubiquitin / ribosomal protein S27a fusion pr... 110 1e-24
xla:444494 rps27a, MGC81889; ribosomal protein S27a; K02977 sm... 95.9 3e-20
hsa:6233 RPS27A, CEP80, UBA80, UBCEP1, UBCEP80; ribosomal prot... 95.9 3e-20
ath:AT1G23410 ubiquitin extension protein, putative / 40S ribo... 95.1 4e-20
mmu:78294 Rps27a, 0610006J14Rik, Uba52, Ubb, Ubc; ribosomal pr... 94.4 9e-20
mmu:667035 Gm8430, EG667035; predicted pseudogene 8430 94.0
dre:337810 rps27a, MGC66168, hm:zeh0386, ik:tdsubc_1f2, tdsubc... 93.6 1e-19
ath:AT2G47110 UBQ6; UBQ6; protein binding; K02977 small subuni... 87.8 7e-18
ath:AT3G62250 UBQ5; UBQ5 (ubiquitin 5); protein binding / stru... 87.4 9e-18
cpv:cgd4_3080 ribosomal protein S27a, ubiquitin plus zincribbo... 78.6 4e-15
tpv:TP04_0752 hypothetical protein 74.3 9e-14
sce:YLR167W RPS31, RPS37, UBI3; Rps31p; K02977 small subunit r... 73.9 1e-13
bbo:BBOV_III008500 17.m07744; ribosomal protein S27a family pr... 71.6 5e-13
pfa:PF14_0027 40S ribosomal protein S31/UBI, putative 57.4 1e-08
cel:K08C9.7 hypothetical protein; K02977 small subunit ribosom... 55.5 4e-08
cel:F34H10.1 hypothetical protein; K02977 small subunit riboso... 54.3 9e-08
hsa:100128355 similar to hCG1790904 40.4 0.001
pfa:PF07_0121 60S ribosomal subunit export protein, putative; ... 28.1 6.3
> cel:H06I04.4 ubl-1; UBiquitin-Like family member (ubl-1); K02977
small subunit ribosomal protein S27Ae
Length=163
Score = 116 bits (290), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 58/97 (59%), Positives = 74/97 (76%), Gaps = 0/97 (0%)
Query 2 EAGVEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRL 61
+ G++ E+T++ +LELLG KKRK+K YT PKK K K KKVKLAVLK+YK+DEN +TRL
Sbjct 52 DCGIDSEATIYVNLELLGGAKKRKKKVYTTPKKNKRKPKKVKLAVLKYYKIDENGKITRL 111
Query 62 RKECPSKTCGSGVFMAQHENRNYCGRCGITYILQSGN 98
RKEC +CG GVFMAQH NR+YCGRC T ++ +
Sbjct 112 RKECQQPSCGGGVFMAQHANRHYCGRCHDTLVVDTAT 148
> tgo:TGME49_045620 ubiquitin / ribosomal protein S27a fusion
protein, putative ; K02977 small subunit ribosomal protein S27Ae
Length=154
Score = 110 bits (275), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 77/95 (81%), Positives = 86/95 (90%), Gaps = 0/95 (0%)
Query 2 EAGVEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRL 61
E GVEEE T++QSLELLGAGKKRK+KTYTKPKKQKHK KKVKLAVLKFYKVD N+ VTRL
Sbjct 58 ELGVEEECTLYQSLELLGAGKKRKKKTYTKPKKQKHKKKKVKLAVLKFYKVDGNDKVTRL 117
Query 62 RKECPSKTCGSGVFMAQHENRNYCGRCGITYILQS 96
RKECP +TCG+GVFMA H+NR YCGRCG+TYIL +
Sbjct 118 RKECPRETCGAGVFMAAHKNRTYCGRCGLTYILNA 152
> xla:444494 rps27a, MGC81889; ribosomal protein S27a; K02977
small subunit ribosomal protein S27Ae
Length=156
Score = 95.9 bits (237), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 56/88 (63%), Positives = 71/88 (80%), Gaps = 0/88 (0%)
Query 5 VEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRLRKE 64
+++EST+ L L G KKRK+K+YT PKK KHK KKVKLAVLK+YKVDEN ++RLR+E
Sbjct 61 IQKESTLHLVLRLRGGAKKRKKKSYTTPKKNKHKRKKVKLAVLKYYKVDENGKISRLRRE 120
Query 65 CPSKTCGSGVFMAQHENRNYCGRCGITY 92
CPS CG+GVFMA H +R+YCG+C +TY
Sbjct 121 CPSDECGAGVFMASHFDRHYCGKCCLTY 148
> hsa:6233 RPS27A, CEP80, UBA80, UBCEP1, UBCEP80; ribosomal protein
S27a; K02977 small subunit ribosomal protein S27Ae
Length=156
Score = 95.9 bits (237), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 56/88 (63%), Positives = 71/88 (80%), Gaps = 0/88 (0%)
Query 5 VEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRLRKE 64
+++EST+ L L G KKRK+K+YT PKK KHK KKVKLAVLK+YKVDEN ++RLR+E
Sbjct 61 IQKESTLHLVLRLRGGAKKRKKKSYTTPKKNKHKRKKVKLAVLKYYKVDENGKISRLRRE 120
Query 65 CPSKTCGSGVFMAQHENRNYCGRCGITY 92
CPS CG+GVFMA H +R+YCG+C +TY
Sbjct 121 CPSDECGAGVFMASHFDRHYCGKCCLTY 148
> ath:AT1G23410 ubiquitin extension protein, putative / 40S ribosomal
protein S27A (RPS27aA); K02977 small subunit ribosomal
protein S27Ae
Length=156
Score = 95.1 bits (235), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 56/94 (59%), Positives = 72/94 (76%), Gaps = 0/94 (0%)
Query 5 VEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRLRKE 64
+++EST+ L L G KKRK+KTYTKPKK KH +KKVKLAVL+FYKVD + V RL+KE
Sbjct 61 IQKESTLHLVLRLRGGAKKRKKKTYTKPKKIKHTHKKVKLAVLQFYKVDGSGKVQRLKKE 120
Query 65 CPSKTCGSGVFMAQHENRNYCGRCGITYILQSGN 98
CPS +CG G FMA H +R+YCG+CG TY+ + +
Sbjct 121 CPSVSCGPGTFMASHFDRHYCGKCGTTYVFKKAD 154
> mmu:78294 Rps27a, 0610006J14Rik, Uba52, Ubb, Ubc; ribosomal
protein S27A; K02977 small subunit ribosomal protein S27Ae
Length=156
Score = 94.4 bits (233), Expect = 9e-20, Method: Compositional matrix adjust.
Identities = 55/88 (62%), Positives = 70/88 (79%), Gaps = 0/88 (0%)
Query 5 VEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRLRKE 64
+++EST+ L L G KKRK+K+YT PKK KHK KKVKLAVLK+YKVDEN ++RLR+E
Sbjct 61 IQKESTLHLVLRLRGGAKKRKKKSYTTPKKNKHKRKKVKLAVLKYYKVDENGKISRLRRE 120
Query 65 CPSKTCGSGVFMAQHENRNYCGRCGITY 92
CPS CG+GVFM H +R+YCG+C +TY
Sbjct 121 CPSDECGAGVFMGSHFDRHYCGKCCLTY 148
> mmu:667035 Gm8430, EG667035; predicted pseudogene 8430
Length=156
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 55/88 (62%), Positives = 70/88 (79%), Gaps = 0/88 (0%)
Query 5 VEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRLRKE 64
+++EST+ L L G KKRK+K+YT PKK KHK KKVKLAVLK+YKVDEN ++RLR+E
Sbjct 61 IQKESTLHLVLRLRGGAKKRKKKSYTTPKKNKHKRKKVKLAVLKYYKVDENGKISRLRRE 120
Query 65 CPSKTCGSGVFMAQHENRNYCGRCGITY 92
CPS CG+GVFM H +R+YCG+C +TY
Sbjct 121 CPSDECGAGVFMGSHFDRHYCGKCCLTY 148
> dre:337810 rps27a, MGC66168, hm:zeh0386, ik:tdsubc_1f2, tdsubc_1f2,
xx:tdsubc_1f2, zgc:66168; ribosomal protein S27a; K02977
small subunit ribosomal protein S27Ae
Length=156
Score = 93.6 bits (231), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 55/88 (62%), Positives = 70/88 (79%), Gaps = 0/88 (0%)
Query 5 VEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRLRKE 64
+++EST+ L L G KKRK+K+YT PKK KHK KKVKLAVLK+YKVDEN + RLR+E
Sbjct 61 IQKESTLHLVLRLRGGAKKRKKKSYTTPKKNKHKRKKVKLAVLKYYKVDENGKIHRLRRE 120
Query 65 CPSKTCGSGVFMAQHENRNYCGRCGITY 92
CP+ CG+GVFMA H +R+YCG+C +TY
Sbjct 121 CPADECGAGVFMASHFDRHYCGKCCLTY 148
> ath:AT2G47110 UBQ6; UBQ6; protein binding; K02977 small subunit
ribosomal protein S27Ae
Length=157
Score = 87.8 bits (216), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 59/91 (64%), Positives = 74/91 (81%), Gaps = 0/91 (0%)
Query 5 VEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRLRKE 64
+++EST+ L L G KKRK+KTYTKPKK KHK+KKVKLAVL+FYKVD + V RLRKE
Sbjct 61 IQKESTLHLVLRLRGGAKKRKKKTYTKPKKIKHKHKKVKLAVLQFYKVDGSGKVQRLRKE 120
Query 65 CPSKTCGSGVFMAQHENRNYCGRCGITYILQ 95
CP+ TCG+G FMA H +R+YCG+CG+TY+ Q
Sbjct 121 CPNATCGAGTFMASHFDRHYCGKCGLTYVYQ 151
> ath:AT3G62250 UBQ5; UBQ5 (ubiquitin 5); protein binding / structural
constituent of ribosome; K02977 small subunit ribosomal
protein S27Ae
Length=157
Score = 87.4 bits (215), Expect = 9e-18, Method: Compositional matrix adjust.
Identities = 58/91 (63%), Positives = 74/91 (81%), Gaps = 0/91 (0%)
Query 5 VEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRLRKE 64
+++EST+ L L G KKRK+KTYTKPKK KHK+KKVKLAVL+FYK+D + V RLRKE
Sbjct 61 IQKESTLHLVLRLRGGAKKRKKKTYTKPKKIKHKHKKVKLAVLQFYKIDGSGKVQRLRKE 120
Query 65 CPSKTCGSGVFMAQHENRNYCGRCGITYILQ 95
CP+ TCG+G FMA H +R+YCG+CG+TY+ Q
Sbjct 121 CPNATCGAGTFMASHFDRHYCGKCGLTYVYQ 151
> cpv:cgd4_3080 ribosomal protein S27a, ubiquitin plus zincribbon,
UB13p ; K02977 small subunit ribosomal protein S27Ae
Length=159
Score = 78.6 bits (192), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 59/98 (60%), Positives = 77/98 (78%), Gaps = 2/98 (2%)
Query 1 EEAGVEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTR 60
E+AG+ + ST++ S +LG G K+K+K +TKPKK KHK KKVKLAVLK+YKVD + V +
Sbjct 60 EQAGISDYSTLYVSEAMLG-GAKKKKKNFTKPKKIKHKKKKVKLAVLKYYKVD-GDKVVK 117
Query 61 LRKECPSKTCGSGVFMAQHENRNYCGRCGITYILQSGN 98
LR+ECP++TCG+GVFMAQH NR CGRCG+TY G+
Sbjct 118 LRRECPAETCGAGVFMAQHFNRTSCGRCGLTYFPSKGD 155
> tpv:TP04_0752 hypothetical protein
Length=162
Score = 74.3 bits (181), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 58/93 (62%), Positives = 67/93 (72%), Gaps = 1/93 (1%)
Query 1 EEAGVEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTR 60
E G E ++ LELLG GKKRK+K YT PKK KHK KKVKLAVLK+YKVD + V R
Sbjct 66 ELLGSESTISLDHHLELLGGGKKRKKKQYTTPKKVKHKKKKVKLAVLKYYKVD-GDQVVR 124
Query 61 LRKECPSKTCGSGVFMAQHENRNYCGRCGITYI 93
L K+CP + CG GVFMA H NR YCGRC +TY+
Sbjct 125 LLKDCPGENCGRGVFMAAHNNRTYCGRCQLTYL 157
> sce:YLR167W RPS31, RPS37, UBI3; Rps31p; K02977 small subunit
ribosomal protein S27Ae
Length=152
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 52/88 (59%), Positives = 67/88 (76%), Gaps = 0/88 (0%)
Query 5 VEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRLRKE 64
+++EST+ L L G GKKRK+K YT PKK KHK+KKVKLAVL +YKVD VT+LR+E
Sbjct 61 IQKESTLHLVLRLRGGGKKRKKKVYTTPKKIKHKHKKVKLAVLSYYKVDAEGKVTKLRRE 120
Query 65 CPSKTCGSGVFMAQHENRNYCGRCGITY 92
C + TCG+GVF+A H++R YCG+C Y
Sbjct 121 CSNPTCGAGVFLANHKDRLYCGKCHSVY 148
> bbo:BBOV_III008500 17.m07744; ribosomal protein S27a family
protein; K02977 small subunit ribosomal protein S27Ae
Length=156
Score = 71.6 bits (174), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 55/91 (60%), Positives = 67/91 (73%), Gaps = 1/91 (1%)
Query 3 AGVEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRLR 62
A +E + Q +++ G GKKRK+K +T PKK KHK KKVKL VLK+YKV E + V RL
Sbjct 63 ASDDEIIELVQHIDVCGGGKKRKKKQHTTPKKIKHKKKKVKLNVLKYYKV-EGDKVVRLL 121
Query 63 KECPSKTCGSGVFMAQHENRNYCGRCGITYI 93
KEC S TCG GVFMAQH +R+YCGRCG TY+
Sbjct 122 KECDSSTCGRGVFMAQHHDRDYCGRCGTTYL 152
> pfa:PF14_0027 40S ribosomal protein S31/UBI, putative
Length=149
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 25/50 (50%), Positives = 35/50 (70%), Gaps = 2/50 (4%)
Query 44 LAVLKFYKVDENNNVTRLRKECPSKTCGSGVFMAQHENRNYCGRCGITYI 93
LAVLKFYKV ++ V RL+++C + C G MA H +R+YCGRC +T +
Sbjct 100 LAVLKFYKVGDDGKVFRLKRQCDN--CAPGTLMASHFDRDYCGRCHLTIM 147
> cel:K08C9.7 hypothetical protein; K02977 small subunit ribosomal
protein S27Ae
Length=154
Score = 55.5 bits (132), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 43/77 (55%), Gaps = 0/77 (0%)
Query 2 EAGVEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRL 61
+ G++ +T++ + ELLG KKRK+K Y PKK +K K++K+DEN ++RL
Sbjct 65 DCGIKAAATIYVNCELLGGAKKRKKKVYIIPKKNNISQRKSSSPSPKYFKIDENGKISRL 124
Query 62 RKECPSKTCGSGVFMAQ 78
P+ + FM Q
Sbjct 125 PTGIPTVSIQREAFMTQ 141
> cel:F34H10.1 hypothetical protein; K02977 small subunit ribosomal
protein S27Ae
Length=142
Score = 54.3 bits (129), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 43/77 (55%), Gaps = 0/77 (0%)
Query 2 EAGVEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRL 61
+ G++ +T++ + ELLG KKRK+K YT PKK +K K++K+DEN ++RL
Sbjct 53 DCGIKAAATIYVNCELLGGAKKRKKKVYTIPKKNNISQRKSSSPSPKYFKIDENGKISRL 112
Query 62 RKECPSKTCGSGVFMAQ 78
+ + FM Q
Sbjct 113 PTGILTASIQREAFMTQ 129
> hsa:100128355 similar to hCG1790904
Length=334
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 32/60 (53%), Positives = 41/60 (68%), Gaps = 1/60 (1%)
Query 2 EAGVEEESTVFQSLELLGAGKKRKRKTYTKPKKQKHKNKKVKLAVLKFYKVDENNNVTRL 61
+ +++E T+ L L G KKRK K+Y PKK KHK KKVKL VLK YKVDEN+ ++ L
Sbjct 126 DCNIQKEPTLHLLLRLCGVAKKRK-KSYPTPKKNKHKRKKVKLDVLKCYKVDENDKISAL 184
> pfa:PF07_0121 60S ribosomal subunit export protein, putative;
K07562 nonsense-mediated mRNA decay protein 3
Length=888
Score = 28.1 bits (61), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 36/79 (45%), Gaps = 3/79 (3%)
Query 24 RKRKTYTKPKKQKHKNKKVKLAVLKFYKV---DENNNVTRLRKECPSKTCGSGVFMAQHE 80
+ YT K K ++KVKL L + V DE+N T L K C +K G ++ ++
Sbjct 751 KNSDIYTDGKSAKSSSRKVKLDKLVYAFVELYDESNGTTILTKTCNAKHLRPGDYVNAYD 810
Query 81 NRNYCGRCGITYILQSGNN 99
R + I L+ +N
Sbjct 811 LRKHSFDNEINLFLEKNDN 829
Lambda K H
0.313 0.130 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2046143372
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40