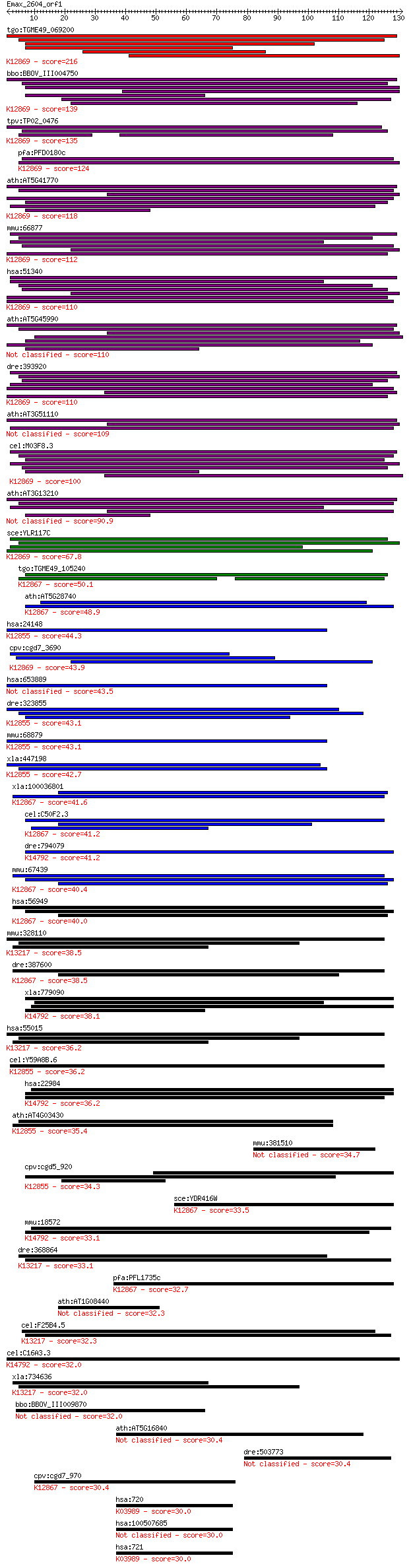
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2604_orf1
Length=130
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_069200 crooked neck-like protein 1, putative ; K128... 216 2e-56
bbo:BBOV_III004750 17.m07426; tetratricopeptide repeat domain ... 139 3e-33
tpv:TP02_0476 crooked neck protein; K12869 crooked neck 135 4e-32
pfa:PFD0180c CGI-201 protein, short form; K12869 crooked neck 124 6e-29
ath:AT5G41770 crooked neck protein, putative / cell cycle prot... 118 4e-27
mmu:66877 Crnkl1, 1200013P10Rik, 5730590A01Rik, C80326, crn; C... 112 3e-25
hsa:51340 CRNKL1, CLF, CRN, Clf1, HCRN, MSTP021, SYF3; crooked... 110 1e-24
ath:AT5G45990 crooked neck protein, putative / cell cycle prot... 110 1e-24
dre:393920 crnkl1, MGC55327, zgc:55327; crooked neck pre-mRNA ... 110 1e-24
ath:AT3G51110 crooked neck protein, putative / cell cycle prot... 109 2e-24
cel:M03F8.3 hypothetical protein; K12869 crooked neck 100 2e-21
ath:AT3G13210 crooked neck protein, putative / cell cycle prot... 90.9 1e-18
sce:YLR117C CLF1, NTC77, SYF3; Clf1p; K12869 crooked neck 67.8 7e-12
tgo:TGME49_105240 XPA-binding protein, putative ; K12867 pre-m... 50.1 2e-06
ath:AT5G28740 transcription-coupled DNA repair protein-related... 48.9 4e-06
hsa:24148 PRPF6, ANT-1, C20orf14, Prp6, TOM, U5-102K, hPrp6; P... 44.3 9e-05
cpv:cgd7_3690 crooked neck protein HAT repeats ; K12869 crooke... 43.9 1e-04
hsa:653889 pre-mRNA-processing factor 6-like 43.5 2e-04
dre:323855 c20orf14, fc12b02, prpf6, wu:fa05f07, wu:fc12b02, z... 43.1 2e-04
mmu:68879 Prpf6, 1190003A07Rik, 2610031L17Rik, ANT-1, MGC11655... 43.1 2e-04
xla:447198 prpf6, MGC80263; PRP6 pre-mRNA processing factor 6 ... 42.7 3e-04
xla:100036801 xab2; XPA binding protein 2; K12867 pre-mRNA-spl... 41.6 6e-04
cel:C50F2.3 hypothetical protein; K12867 pre-mRNA-splicing fac... 41.2 8e-04
dre:794079 pdcd11, MGC162501, cb680, im:7148359, sb:cb680, wu:... 41.2 8e-04
mmu:67439 Xab2, 0610041O14Rik, AV025587; XPA binding protein 2... 40.4 0.002
hsa:56949 XAB2, DKFZp762C1015, HCNP, HCRN, NTC90, SYF1; XPA bi... 40.0 0.002
mmu:328110 Prpf39, FLJ11128, MGC37077, Srcs1; PRP39 pre-mRNA p... 38.5 0.005
dre:387600 xab2, MGC198247, zgc:63498, zgc:63949; XPA binding ... 38.5 0.005
xla:779090 pdcd11, alg-4, alg4, nfbp, rrp5; programmed cell de... 38.1 0.007
hsa:55015 PRPF39, FLJ11128, FLJ20666, FLJ45460, MGC149842, MGC... 36.2 0.024
cel:Y59A8B.6 hypothetical protein; K12855 pre-mRNA-processing ... 36.2 0.025
hsa:22984 PDCD11, ALG-4, ALG4, KIAA0185, NFBP, RRP5; programme... 36.2 0.029
ath:AT4G03430 EMB2770 (EMBRYO DEFECTIVE 2770); RNA splicing fa... 35.4 0.046
mmu:381510 Dpy19l4, Gm1023, Narg3; dpy-19-like 4 (C. elegans) 34.7 0.075
cpv:cgd5_920 Pre-mRNA splicing factor Pro1/Prp6. HAT repeat pr... 34.3 0.10
sce:YDR416W SYF1, NTC90; Member of the NineTeen Complex (NTC) ... 33.5 0.18
mmu:18572 Pdcd11, 1110021I22Rik, ALG-4, Pdcd7, mKIAA0185; prog... 33.1 0.22
dre:368864 prpf39, fa10d07, si:dz261o22.3, wu:fa10d07; PRP39 p... 33.1 0.24
pfa:PFL1735c RNA-processing protein, putative; K12867 pre-mRNA... 32.7 0.27
ath:AT1G08440 hypothetical protein 32.3 0.34
cel:F25B4.5 hypothetical protein; K13217 pre-mRNA-processing f... 32.3 0.42
cel:C16A3.3 let-716; LEThal family member (let-716); K14792 rR... 32.0 0.44
xla:734636 prpf39.1, MGC115228, prpf39; PRP39 pre-mRNA process... 32.0 0.45
bbo:BBOV_III009870 17.m07855; hypothetical protein 32.0 0.55
ath:AT5G16840 BPA1; BPA1 (BINDINGPARTNEROFACD11 1); nucleic ac... 30.4 1.4
dre:503773 MGC158780, id:ibd5127, wu:fa56d01, zgc:158780; zgc:... 30.4 1.6
cpv:cgd7_970 Syf1p. protein with 8 HAT domains ; K12867 pre-mR... 30.4 1.6
hsa:720 C4A, C4, C4A2, C4A3, C4A4, C4A6, C4S, CO4, CPAMD2, MGC... 30.0 1.8
hsa:100507685 complement C4-B-like 30.0 1.8
hsa:721 C4B, C4B1, C4B12, C4B2, C4B3, C4B5, C4F, CH, CO4, CPAM... 30.0 1.8
> tgo:TGME49_069200 crooked neck-like protein 1, putative ; K12869
crooked neck
Length=794
Score = 216 bits (549), Expect = 2e-56, Method: Compositional matrix adjust.
Identities = 96/128 (75%), Positives = 120/128 (93%), Gaps = 0/128 (0%)
Query 1 NPSDKGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGY 60
NPSDKGW++YIHFEERC+EL+RAR++FERYLSNRPSQE+FLRFCKFEE+H+ IPRARAG+
Sbjct 303 NPSDKGWMLYIHFEERCKELDRARKVFERYLSNRPSQESFLRFCKFEERHRQIPRARAGF 362
Query 61 EKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQ 120
EKAIELLPED+L+EHF++KFA FEERQ+E ERAK IY+ AL+++P+G++D LY KYV+FQ
Sbjct 363 EKAIELLPEDMLDEHFFLKFAQFEERQRETERAKVIYQQALEQLPKGESDLLYEKYVTFQ 422
Query 121 KQFGEHEG 128
KQFG+ EG
Sbjct 423 KQFGDKEG 430
Score = 51.6 bits (122), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 35/120 (29%), Positives = 58/120 (48%), Gaps = 3/120 (2%)
Query 5 KGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGYEKAI 64
K W +Y FE R R+L++AR IF R ++ + F+ + + E + I R R Y K I
Sbjct 537 KIWSLYASFEVRQRDLDKARLIFGRAIAECGKPKIFVAYAQLELRLGCIDRCRKIYAKFI 596
Query 65 ELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFG 124
EL P N ++ E +E+ RA+A+ E A+ + L+ Y+ + +G
Sbjct 597 ELHP---FNPRAWIAMIDLEVLAEEQARARALCELAIGMEEMDTPELLWKAYIDMEVGWG 653
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 29/107 (27%), Positives = 57/107 (53%), Gaps = 12/107 (11%)
Query 7 WLVYIHFEERCRELERARRIFERYLSNRPSQE----------AFLRFCKFEE-KHKNIPR 55
W+ YI EE ++++ R ++ER L+N P ++ + FEE + K++ R
Sbjct 457 WIDYIRLEESRGDIDKIRNVYERALANVPPVLEKRFWKRYVYIWISYALFEELQAKDVER 516
Query 56 ARAGYEKAIELLPEDLLN-EHFYMKFAAFEERQQEKERAKAIYEAAL 101
R Y K +E++P + + +A+FE RQ++ ++A+ I+ A+
Sbjct 517 CRQVYVKMLEVIPHKKFSFAKIWSLYASFEVRQRDLDKARLIFGRAI 563
Score = 35.0 bits (79), Expect = 0.051, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 35/69 (50%), Gaps = 1/69 (1%)
Query 7 WLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEK-HKNIPRARAGYEKAIE 65
W YI E ++RAR ++ER L + F F FE + +++P AR E+ IE
Sbjct 642 WKAYIDMEVGWGAVDRARSLYERLLEKTQHVKVFKSFADFEWRIVESLPNARKVIERGIE 701
Query 66 LLPEDLLNE 74
+ E+ +E
Sbjct 702 VCKENSWDE 710
Score = 30.8 bits (68), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 33/61 (54%), Gaps = 4/61 (6%)
Query 26 IFERYLS-NRPSQEAFLRFCKFEEKHKNIPRARAGYEKAIELLPEDLLNEHFYMKFAAFE 84
+FER L+ + + +L++ + E K+K I R Y++ LLP E F+ K+A E
Sbjct 129 VFERALNVDFQNTTLWLKYIEMESKNKFINSCRNLYDRVCLLLPR---QEQFWFKYAHME 185
Query 85 E 85
E
Sbjct 186 E 186
Score = 28.5 bits (62), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 23/89 (25%), Positives = 40/89 (44%), Gaps = 6/89 (6%)
Query 41 LRFCKFEEKHKNIPRARAGYEKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAA 100
LR C + + AR +E+ +E P D +M + FEER +E +RA+ ++E
Sbjct 279 LRLCS--PGRRALAGARNVFERWMEWNPSD----KGWMLYIHFEERCKELDRARKVFERY 332
Query 101 LQRVPRGQADELYSKYVSFQKQFGEHEGG 129
L P ++ + K+ +Q G
Sbjct 333 LSNRPSQESFLRFCKFEERHRQIPRARAG 361
> bbo:BBOV_III004750 17.m07426; tetratricopeptide repeat domain
containing protein; K12869 crooked neck
Length=665
Score = 139 bits (349), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 65/128 (50%), Positives = 91/128 (71%), Gaps = 0/128 (0%)
Query 1 NPSDKGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGY 60
NP D+ W++YI FEERC EL+R R+IFER+L +RPS +FL+F KFE++ KN P ARA Y
Sbjct 171 NPDDRSWMLYIKFEERCGELDRCRQIFERFLESRPSCASFLKFAKFEQRQKNYPLARAAY 230
Query 61 EKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQ 120
K +E++P +LL E F++KFAAFE +Q A+ +YE L +PR +++LY +VSFQ
Sbjct 231 VKCLEIIPPELLTEEFFLKFAAFETQQGNLSGAEKVYEQGLGILPRESSEQLYRSFVSFQ 290
Query 121 KQFGEHEG 128
KQ + E
Sbjct 291 KQHRDRET 298
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 36/121 (29%), Positives = 64/121 (52%), Gaps = 7/121 (5%)
Query 6 GWLVYIHFEERCRELERARRIFERYLSNRPSQ-EAFLRFCKFEEKHKNIPRARAGYEKAI 64
W+ Y +E ++ RAR +FER L P+ +LR+ + E K+KN+ AR +++ +
Sbjct 75 TWIKYALWEANQQDFRRARSVFERALQVDPNNVNLWLRYIETEMKNKNVNAARNLFDRVV 134
Query 65 ELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFG 124
LLP + F+ K+A FEE A+ ++E ++ P D + Y+ F+++ G
Sbjct 135 SLLPR---VDQFWFKYAHFEELLGNYAGARTVFERWMEWNP---DDRSWMLYIKFEERCG 188
Query 125 E 125
E
Sbjct 189 E 189
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 42/154 (27%), Positives = 68/154 (44%), Gaps = 31/154 (20%)
Query 7 WLVYIHFEERCRELERARRIFERYLSNRPSQEAF-LRFCKFEEKHKNIPRARAGYEKAIE 65
WL YI E + + + AR +F+R +S P + F ++ FEE N AR +E+ +E
Sbjct 110 WLRYIETEMKNKNVNAARNLFDRVVSLLPRVDQFWFKYAHFEELLGNYAGARTVFERWME 169
Query 66 LLPED---LLNEHF--------------------------YMKFAAFEERQQEKERAKAI 96
P+D +L F ++KFA FE+RQ+ A+A
Sbjct 170 WNPDDRSWMLYIKFEERCGELDRCRQIFERFLESRPSCASFLKFAKFEQRQKNYPLARAA 229
Query 97 YEAALQRV-PRGQADELYSKYVSFQKQFGEHEGG 129
Y L+ + P +E + K+ +F+ Q G G
Sbjct 230 YVKCLEIIPPELLTEEFFLKFAAFETQQGNLSGA 263
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 18/91 (19%), Positives = 49/91 (53%), Gaps = 5/91 (5%)
Query 39 AFLRFCKFEEKHKNIPRARAGYEKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYE 98
++++ +E ++ RAR+ +E+A+++ P N + ++++ E + + A+ +++
Sbjct 75 TWIKYALWEANQQDFRRARSVFERALQVDPN---NVNLWLRYIETEMKNKNVNAARNLFD 131
Query 99 AALQRVPRGQADELYSKYVSFQKQFGEHEGG 129
+ +PR D+ + KY F++ G + G
Sbjct 132 RVVSLLPR--VDQFWFKYAHFEELLGNYAGA 160
Score = 33.1 bits (74), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 20/59 (33%), Positives = 28/59 (47%), Gaps = 0/59 (0%)
Query 7 WLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGYEKAIE 65
W YI E ++ R I+ER L + FL +C+FE ARA E+A+E
Sbjct 522 WNRYIEIEREWQQYAHVRNIYERLLLKTTHIKVFLSYCEFEFTSGFPDNARAIAERALE 580
Score = 32.7 bits (73), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 22/108 (20%), Positives = 49/108 (45%), Gaps = 3/108 (2%)
Query 19 ELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGYEKAIELLPEDLLNEHFYM 78
+L+ R+ F L + F + + E K N+ R R + K IE P ++
Sbjct 431 DLDSMRKTFGLGLGQCKKPKLFETYAQIELKLGNLDRCRHIHAKYIETWP---FKPESWL 487
Query 79 KFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFGEH 126
F E E++R + + EAA+ + ++++Y+ ++++ ++
Sbjct 488 SFIELELMLNERKRVRGLCEAAIAMDQMDMPETVWNRYIEIEREWQQY 535
Score = 29.3 bits (64), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 25/105 (23%), Positives = 48/105 (45%), Gaps = 13/105 (12%)
Query 22 RARRIFERYLSNRPSQE----------AFLRFCKFEE-KHKNIPRARAGYEKAIELLPED 70
R ++ER +SN P + ++ + F E + + RA A Y KA+++LP+D
Sbjct 355 RVCELYERAISNLPQVDDRRLWRRYSYLWVGYAIFSELTLQQLDRAVAVYRKALQVLPKD 414
Query 71 LLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSK 115
FY+ A RQ + + + + L + + + E Y++
Sbjct 415 F--AKFYILLAELYLRQGDLDSMRKTFGLGLGQCKKPKLFETYAQ 457
> tpv:TP02_0476 crooked neck protein; K12869 crooked neck
Length=657
Score = 135 bits (339), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 60/123 (48%), Positives = 88/123 (71%), Gaps = 0/123 (0%)
Query 1 NPSDKGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGY 60
NP DK W++YI FEERC E++R R IF RY+ NRPS +FL+ KFEEK+K RAR+ +
Sbjct 171 NPEDKAWMLYIKFEERCGEVDRCRSIFNRYIENRPSCMSFLKLVKFEEKYKKTSRARSAF 230
Query 61 EKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQ 120
K +E+L +LL+E F++KFA FE+R E A ++YE L+ + + ++++LY ++SFQ
Sbjct 231 VKCVEVLDPELLDEDFFIKFANFEQRHNNIEGANSVYEQGLKLLDKSKSEKLYDSFISFQ 290
Query 121 KQF 123
KQF
Sbjct 291 KQF 293
Score = 61.2 bits (147), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 41/121 (33%), Positives = 67/121 (55%), Gaps = 7/121 (5%)
Query 6 GWLVYIHFEERCRELERARRIFERYLSNRPSQEA-FLRFCKFEEKHKNIPRARAGYEKAI 64
W+ Y +E +E RAR IFER L P+ + +LR+ + E K+KNI AR +++ +
Sbjct 75 TWIKYAVWEANQQEFRRARSIFERALLVDPNNPSLWLRYIETEMKNKNINSARNLFDRVV 134
Query 65 ELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFG 124
LLP + F+ K+A FEE A++IYE ++ P +A + Y+ F+++ G
Sbjct 135 CLLPRI---DQFWFKYAHFEELLGNYAGARSIYERWMEWNPEDKA---WMLYIKFEERCG 188
Query 125 E 125
E
Sbjct 189 E 189
Score = 31.6 bits (70), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 39/77 (50%), Gaps = 7/77 (9%)
Query 38 EAFLRFCKFEEKHKNIPRARAGYEKAIELLPED----LLNEHFYMK--FAAFEERQQE-K 90
E L+ C ++ R YE+AI LP+D L + Y+ +A F E Q + K
Sbjct 336 ENMLKTCSDDKLGAQKDRIVQVYERAIANLPKDNNRKLWRRYSYLWIFYAFFSELQLDSK 395
Query 91 ERAKAIYEAALQRVPRG 107
ERA+ IY +LQ +PR
Sbjct 396 ERAEEIYLKSLQILPRD 412
Score = 27.7 bits (60), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 11/24 (45%), Positives = 15/24 (62%), Gaps = 0/24 (0%)
Query 5 KGWLVYIHFEERCRELERARRIFE 28
K WL YI+FE E+ R R++ E
Sbjct 482 KSWLAYINFELLLNEINRVRKLCE 505
> pfa:PFD0180c CGI-201 protein, short form; K12869 crooked neck
Length=780
Score = 124 bits (312), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 59/122 (48%), Positives = 83/122 (68%), Gaps = 0/122 (0%)
Query 6 GWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGYEKAIE 65
+L YI+FEERC E+ + R IFER + + P E F +F KFE+K+KNI RARA YEK IE
Sbjct 174 SFLCYIYFEERCNEINKCREIFERLIVSIPKLECFYKFIKFEKKYKNIVRARAAYEKCIE 233
Query 66 LLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFGE 125
LLP ++E+FY+ F FEE Q E ER K IY AL+ +P+ +++ LY ++ FQK++
Sbjct 234 LLPSCYIDENFYIHFCNFEEEQNEYERCKKIYIEALKILPKNKSELLYKNFLQFQKKYAN 293
Query 126 HE 127
+
Sbjct 294 KD 295
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 36/125 (28%), Positives = 58/125 (46%), Gaps = 3/125 (2%)
Query 5 KGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGYEKAI 64
K +++Y FE R + +AR IF L P+++ F +FC+FE K NI R Y K +
Sbjct 410 KIFILYATFELRQLNVNKARSIFNNALQTIPNEKIFEKFCEFELKLGNIRECRNVYAKYV 469
Query 65 ELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFG 124
E P N ++ FE E ERA+ I E A+ + ++ Y+ +
Sbjct 470 EAFP---FNSKAWISMINFELSLDEVERARQIAEIAINLDDMKLPELIWKNYIDMEINLQ 526
Query 125 EHEGG 129
E++
Sbjct 527 EYDNA 531
> ath:AT5G41770 crooked neck protein, putative / cell cycle protein,
putative; K12869 crooked neck
Length=705
Score = 118 bits (296), Expect = 4e-27, Method: Compositional matrix adjust.
Identities = 59/128 (46%), Positives = 88/128 (68%), Gaps = 0/128 (0%)
Query 1 NPSDKGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGY 60
+P +GWL +I FE R E+ERAR I+ER++ P A++R+ KFE K + R R+ Y
Sbjct 190 SPDQQGWLSFIKFELRYNEIERARTIYERFVLCHPKVSAYIRYAKFEMKGGEVARCRSVY 249
Query 61 EKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQ 120
E+A E L +D E ++ FA FEER +E ERA+ IY+ AL +P+G+A++LY K+V+F+
Sbjct 250 ERATEKLADDEEAEILFVAFAEFEERCKEVERARFIYKFALDHIPKGRAEDLYRKFVAFE 309
Query 121 KQFGEHEG 128
KQ+G+ EG
Sbjct 310 KQYGDKEG 317
Score = 58.2 bits (139), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 39/123 (31%), Positives = 61/123 (49%), Gaps = 3/123 (2%)
Query 5 KGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGYEKAI 64
K WL+ FE R L AR+I + P + F ++ + E + N+ R R YE+ +
Sbjct 424 KIWLLAAQFEIRQLNLTGARQILGNAIGKAPKDKIFKKYIEIELQLGNMDRCRKLYERYL 483
Query 65 ELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFG 124
E PE N + + K+A E E ERA+AI+E A+ + + L+ Y+ F+ G
Sbjct 484 EWSPE---NCYAWSKYAELERSLVETERARAIFELAISQPALDMPELLWKAYIDFEISEG 540
Query 125 EHE 127
E E
Sbjct 541 ELE 543
Score = 50.8 bits (120), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 27/96 (28%), Positives = 55/96 (57%), Gaps = 5/96 (5%)
Query 34 RPSQEAFLRFCKFEEKHKNIPRARAGYEKAIELLPEDLLNEHFYMKFAAFEERQQEKERA 93
R + + ++++ ++EE K+ RAR+ +E+AIE D N ++K+A FE + + A
Sbjct 89 RWNIQVWVKYAQWEESQKDYARARSVWERAIE---GDYRNHTLWLKYAEFEMKNKFVNSA 145
Query 94 KAIYEAALQRVPRGQADELYSKYVSFQKQFGEHEGG 129
+ +++ A+ +PR D+L+ KY+ ++ G G
Sbjct 146 RNVWDRAVTLLPR--VDQLWYKYIHMEEILGNIAGA 179
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 37/140 (26%), Positives = 67/140 (47%), Gaps = 16/140 (11%)
Query 1 NPSD-KGWLVYIHFEERCRELERARRIFERYLSNRPSQE----------AFLRFCKFEE- 48
+PS+ W Y+ EE +R R I+ER ++N P E ++ + FEE
Sbjct 337 SPSNYDSWFDYVRLEESVGNKDRIREIYERAIANVPPAEEKRYWQRYIYLWINYALFEEI 396
Query 49 KHKNIPRARAGYEKAIELLPEDLLN-EHFYMKFAAFEERQQEKERAKAIYEAALQRVPRG 107
+ ++I R R Y + ++L+P + ++ A FE RQ A+ I A+ + P+
Sbjct 397 ETEDIERTRDVYRECLKLIPHSKFSFAKIWLLAAQFEIRQLNLTGARQILGNAIGKAPK- 455
Query 108 QADELYSKYVSFQKQFGEHE 127
D+++ KY+ + Q G +
Sbjct 456 --DKIFKKYIEIELQLGNMD 473
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 40/150 (26%), Positives = 71/150 (47%), Gaps = 34/150 (22%)
Query 7 WLVYIHFEERCRELERARRIFERYL-SNRPSQEAFLRFCKFEEKHKNIPRARAGYEKAIE 65
W+ Y +EE ++ RAR ++ER + + + +L++ +FE K+K + AR +++A+
Sbjct 95 WVKYAQWEESQKDYARARSVWERAIEGDYRNHTLWLKYAEFEMKNKFVNSARNVWDRAVT 154
Query 66 LLP------------EDLL------------------NEHFYMKFAAFEERQQEKERAKA 95
LLP E++L ++ ++ F FE R E ERA+
Sbjct 155 LLPRVDQLWYKYIHMEEILGNIAGARQIFERWMDWSPDQQGWLSFIKFELRYNEIERART 214
Query 96 IYEAALQRVPRGQADELYSKYVSFQKQFGE 125
IYE + P+ A Y +Y F+ + GE
Sbjct 215 IYERFVLCHPKVSA---YIRYAKFEMKGGE 241
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 39/121 (32%), Positives = 58/121 (47%), Gaps = 8/121 (6%)
Query 2 PSDKGWLVYIHFEERCRELERARRIFERYLSNRPSQ-EAFLRFCKFEEKHKNIPRARAGY 60
P DK + YI E + ++R R+++ERYL P A+ ++ + E RARA +
Sbjct 454 PKDKIFKKYIEIELQLGNMDRCRKLYERYLEWSPENCYAWSKYAELERSLVETERARAIF 513
Query 61 EKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQ 120
E AI P + E + + FE + E ER +A+YE L R Y +VSF
Sbjct 514 ELAIS-QPALDMPELLWKAYIDFEISEGELERTRALYERLLDRTKH------YKVWVSFA 566
Query 121 K 121
K
Sbjct 567 K 567
Score = 32.7 bits (73), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 17/41 (41%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 7 WLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFE 47
W YI FE ELER R ++ER L + ++ F KFE
Sbjct 529 WKAYIDFEISEGELERTRALYERLLDRTKHYKVWVSFAKFE 569
> mmu:66877 Crnkl1, 1200013P10Rik, 5730590A01Rik, C80326, crn;
Crn, crooked neck-like 1 (Drosophila); K12869 crooked neck
Length=690
Score = 112 bits (280), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 53/127 (41%), Positives = 85/127 (66%), Gaps = 0/127 (0%)
Query 2 PSDKGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGYE 61
P ++ W YI+FE R +E+ERAR I+ER++ P+ + ++++ +FEEKH AR YE
Sbjct 180 PEEQAWHSYINFELRYKEVERARTIYERFVLVHPAVKNWIKYARFEEKHAYFAHARKVYE 239
Query 62 KAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQK 121
+A+E ++ ++EH Y+ FA FEE Q+E ER + IY+ AL R+ + +A EL+ Y F+K
Sbjct 240 RAVEFFGDEHMDEHLYVAFAKFEENQKEFERVRVIYKYALDRISKQEAQELFKNYTIFEK 299
Query 122 QFGEHEG 128
+FG+ G
Sbjct 300 KFGDRRG 306
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 37/116 (31%), Positives = 57/116 (49%), Gaps = 3/116 (2%)
Query 5 KGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGYEKAI 64
K WL Y FE R + L ARR + P + F + + E + + R R YEK +
Sbjct 413 KMWLYYAQFEIRQKNLPFARRALGTSIGKCPKNKLFKGYIELELQLREFDRCRKLYEKFL 472
Query 65 ELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQ 120
E PE+ + ++KFA E + ERA+AIYE A+ + + L+ Y+ F+
Sbjct 473 EFGPENCTS---WIKFAELETILGDIERARAIYELAISQPRLDMPEVLWKSYIDFE 525
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 35/104 (33%), Positives = 56/104 (53%), Gaps = 2/104 (1%)
Query 2 PSDKGWLVYIHFEERCRELERARRIFERYLSNRPSQ-EAFLRFCKFEEKHKNIPRARAGY 60
P +K + YI E + RE +R R+++E++L P ++++F + E +I RARA Y
Sbjct 443 PKNKLFKGYIELELQLREFDRCRKLYEKFLEFGPENCTSWIKFAELETILGDIERARAIY 502
Query 61 EKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRV 104
E AI P + E + + FE Q+E ER + +Y LQR
Sbjct 503 ELAISQ-PRLDMPEVLWKSYIDFEIEQEETERTRNLYRQLLQRT 545
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 35/123 (28%), Positives = 68/123 (55%), Gaps = 7/123 (5%)
Query 6 GWLVYIHFEERCRELERARRIFERYLS-NRPSQEAFLRFCKFEEKHKNIPRARAGYEKAI 64
W+ Y +EE +E++RAR I+ER L + + +L++ + E K++ + AR +++AI
Sbjct 83 NWIKYAQWEESLKEIQRARSIYERALDVDYRNITLWLKYAEMEMKNRQVNHARNIWDRAI 142
Query 65 ELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFG 124
LP +N+ F+ K+ EE A+ ++E ++ P QA + Y++F+ ++
Sbjct 143 TTLPR--VNQ-FWYKYTYMEEMLGNVAGARQVFERWMEWQPEEQA---WHSYINFELRYK 196
Query 125 EHE 127
E E
Sbjct 197 EVE 199
Score = 51.2 bits (121), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 30/109 (27%), Positives = 59/109 (54%), Gaps = 6/109 (5%)
Query 22 RARRIFERYL-SNRPSQEAFLRFCKFEEKHKNIPRARAGYEKAIELLPEDLLNEHFYMKF 80
R R+ FE + NR ++++ ++EE K I RAR+ YE+A+++ D N ++K+
Sbjct 65 RKRKTFEDNIRKNRTVISNWIKYAQWEESLKEIQRARSIYERALDV---DYRNITLWLKY 121
Query 81 AAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFGEHEGG 129
A E + ++ A+ I++ A+ +PR ++ + KY ++ G G
Sbjct 122 AEMEMKNRQVNHARNIWDRAITTLPR--VNQFWYKYTYMEEMLGNVAGA 168
Score = 37.0 bits (84), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 31/138 (22%), Positives = 60/138 (43%), Gaps = 16/138 (11%)
Query 1 NPSD-KGWLVYIHFEERCRELERARRIFERYLSNRPSQE----------AFLRFCKFEEK 49
NP + W Y+ E E + R ++ER ++N P + ++ + +EE
Sbjct 326 NPHNYDAWFDYLRLVESDAEADTVREVYERAIANVPPIQEKRHWKRYIYLWVNYALYEEL 385
Query 50 HKNIP-RARAGYEKAIELLPEDLLN-EHFYMKFAAFEERQQEKERAKAIYEAALQRVPRG 107
P R R Y+ ++EL+P ++ +A FE RQ+ A+ ++ + P+
Sbjct 386 EAKDPERTRQVYQASLELIPHKKFTFAKMWLYYAQFEIRQKNLPFARRALGTSIGKCPKN 445
Query 108 QADELYSKYVSFQKQFGE 125
+ L+ Y+ + Q E
Sbjct 446 K---LFKGYIELELQLRE 460
> hsa:51340 CRNKL1, CLF, CRN, Clf1, HCRN, MSTP021, SYF3; crooked
neck pre-mRNA splicing factor-like 1 (Drosophila); K12869
crooked neck
Length=848
Score = 110 bits (275), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 52/127 (40%), Positives = 83/127 (65%), Gaps = 0/127 (0%)
Query 2 PSDKGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGYE 61
P ++ W YI+FE R +E++RAR I+ER++ P + ++++ +FEEKH AR YE
Sbjct 341 PEEQAWHSYINFELRYKEVDRARTIYERFVLVHPDVKNWIKYARFEEKHAYFAHARKVYE 400
Query 62 KAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQK 121
+A+E ++ ++EH Y+ FA FEE Q+E ER + IY+ AL R+ + A EL+ Y F+K
Sbjct 401 RAVEFFGDEHMDEHLYVAFAKFEENQKEFERVRVIYKYALDRISKQDAQELFKNYTIFEK 460
Query 122 QFGEHEG 128
+FG+ G
Sbjct 461 KFGDRRG 467
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 36/104 (34%), Positives = 57/104 (54%), Gaps = 2/104 (1%)
Query 2 PSDKGWLVYIHFEERCRELERARRIFERYLSNRPSQ-EAFLRFCKFEEKHKNIPRARAGY 60
P +K + VYI E + RE +R R+++E++L P ++++F + E +I RARA Y
Sbjct 604 PKNKLFKVYIELELQLREFDRCRKLYEKFLEFGPENCTSWIKFAELETILGDIDRARAIY 663
Query 61 EKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRV 104
E AI P + E + + FE Q+E ER + +Y LQR
Sbjct 664 ELAISQ-PRLDMPEVLWKSYIDFEIEQEETERTRNLYRRLLQRT 706
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 35/116 (30%), Positives = 58/116 (50%), Gaps = 3/116 (2%)
Query 5 KGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGYEKAI 64
K W++Y FE R + L ARR + P + F + + E + + R R YEK +
Sbjct 574 KMWILYAQFEIRQKNLSLARRALGTSIGKCPKNKLFKVYIELELQLREFDRCRKLYEKFL 633
Query 65 ELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQ 120
E PE+ + ++KFA E + +RA+AIYE A+ + + L+ Y+ F+
Sbjct 634 EFGPENCTS---WIKFAELETILGDIDRARAIYELAISQPRLDMPEVLWKSYIDFE 686
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 34/121 (28%), Positives = 67/121 (55%), Gaps = 7/121 (5%)
Query 6 GWLVYIHFEERCRELERARRIFERYLS-NRPSQEAFLRFCKFEEKHKNIPRARAGYEKAI 64
W+ Y +EE +E++RAR I+ER L + + +L++ + E K++ + AR +++AI
Sbjct 244 NWIKYAQWEESLKEIQRARSIYERALDVDYRNITLWLKYAEMEMKNRQVNHARNIWDRAI 303
Query 65 ELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFG 124
LP +N+ F+ K+ EE A+ ++E ++ P QA + Y++F+ ++
Sbjct 304 TTLPR--VNQ-FWYKYTYMEEMLGNVAGARQVFERWMEWQPEEQA---WHSYINFELRYK 357
Query 125 E 125
E
Sbjct 358 E 358
Score = 51.6 bits (122), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 30/109 (27%), Positives = 59/109 (54%), Gaps = 6/109 (5%)
Query 22 RARRIFERYL-SNRPSQEAFLRFCKFEEKHKNIPRARAGYEKAIELLPEDLLNEHFYMKF 80
R R+ FE + NR ++++ ++EE K I RAR+ YE+A+++ D N ++K+
Sbjct 226 RKRKTFEDNIRKNRTVISNWIKYAQWEESLKEIQRARSIYERALDV---DYRNITLWLKY 282
Query 81 AAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFGEHEGG 129
A E + ++ A+ I++ A+ +PR ++ + KY ++ G G
Sbjct 283 AEMEMKNRQVNHARNIWDRAITTLPR--VNQFWYKYTYMEEMLGNVAGA 329
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 32/138 (23%), Positives = 61/138 (44%), Gaps = 16/138 (11%)
Query 1 NPSD-KGWLVYIHFEERCRELERARRIFERYLSNRPSQE----------AFLRFCKFEEK 49
NP + W Y+ E E E R ++ER ++N P + ++ + +EE
Sbjct 487 NPHNYDAWFDYLRLVESDAEAEAVREVYERAIANVPPIQEKRHWKRYIYLWINYALYEEL 546
Query 50 HKNIP-RARAGYEKAIELLPEDLLN-EHFYMKFAAFEERQQEKERAKAIYEAALQRVPRG 107
P R R Y+ ++EL+P ++ +A FE RQ+ A+ ++ + P+
Sbjct 547 EAKDPERTRQVYQASLELIPHKKFTFAKMWILYAQFEIRQKNLSLARRALGTSIGKCPK- 605
Query 108 QADELYSKYVSFQKQFGE 125
++L+ Y+ + Q E
Sbjct 606 --NKLFKVYIELELQLRE 621
Score = 35.8 bits (81), Expect = 0.037, Method: Compositional matrix adjust.
Identities = 37/172 (21%), Positives = 62/172 (36%), Gaps = 45/172 (26%)
Query 1 NPSDKGWLVYIHFEERCRELERARRIFERYL----SNRPSQEAFLRFCKFEEKHKNIPRA 56
+P K W+ Y FEE+ AR+++ER + + ++ F KFEE K R
Sbjct 373 HPDVKNWIKYARFEEKHAYFAHARKVYERAVEFFGDEHMDEHLYVAFAKFEENQKEFERV 432
Query 57 RAGYEKAIELLP------------------------EDLL--------------NEHFY- 77
R Y+ A++ + ED++ N H Y
Sbjct 433 RVIYKYALDRISKQDAQELFKNYTIFEKKFGDRRGIEDIIVSKRRFQYEEEVKANPHNYD 492
Query 78 --MKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFGEHE 127
+ E E E + +YE A+ VP Q + +Y+ + +E
Sbjct 493 AWFDYLRLVESDAEAEAVREVYERAIANVPPIQEKRHWKRYIYLWINYALYE 544
> ath:AT5G45990 crooked neck protein, putative / cell cycle protein,
putative
Length=673
Score = 110 bits (275), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 59/129 (45%), Positives = 85/129 (65%), Gaps = 1/129 (0%)
Query 1 NPSDKGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKN-IPRARAG 59
+P K WL +I FE R E+ERAR I+ER++ P AF+R+ KFE K + AR
Sbjct 176 SPDQKAWLCFIKFELRYNEIERARSIYERFVLCHPKVSAFIRYAKFEMKRGGQVKLAREV 235
Query 60 YEKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSF 119
YE+A++ L D E ++ FA FEER +E ERA+ IY+ AL + +G+A+ELY K+V+F
Sbjct 236 YERAVDKLANDEEAEILFVSFAEFEERCKEVERARFIYKFALDHIRKGRAEELYKKFVAF 295
Query 120 QKQFGEHEG 128
+KQ+G+ EG
Sbjct 296 EKQYGDKEG 304
Score = 58.2 bits (139), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 40/123 (32%), Positives = 61/123 (49%), Gaps = 3/123 (2%)
Query 5 KGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGYEKAI 64
K WL+ +E R L AR+I + P + F ++ + E K NI R R YE+ +
Sbjct 411 KIWLLAAEYEIRQLNLTGARQILGNAIGKAPKVKIFKKYIEMELKLVNIDRCRKLYERFL 470
Query 65 ELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFG 124
E PE N + + +A FE E ERA+AI+E A+ + + L+ Y+ F+ G
Sbjct 471 EWSPE---NCYAWRNYAEFEISLAETERARAIFELAISQPALDMPELLWKTYIDFEISEG 527
Query 125 EHE 127
E E
Sbjct 528 EFE 530
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 24/96 (25%), Positives = 55/96 (57%), Gaps = 5/96 (5%)
Query 34 RPSQEAFLRFCKFEEKHKNIPRARAGYEKAIELLPEDLLNEHFYMKFAAFEERQQEKERA 93
R + + ++++ K+EE + RAR+ +E+A+E + N ++K+A FE + + A
Sbjct 75 RWNIQVWVKYAKWEESQMDYARARSVWERALE---GEYRNHTLWVKYAEFEMKNKFVNNA 131
Query 94 KAIYEAALQRVPRGQADELYSKYVSFQKQFGEHEGG 129
+ +++ ++ +PR D+L+ KY+ +++ G G
Sbjct 132 RNVWDRSVTLLPR--VDQLWEKYIYMEEKLGNVTGA 165
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 34/122 (27%), Positives = 58/122 (47%), Gaps = 5/122 (4%)
Query 10 YIHFEERCRELERARRIFERYLSNRPSQ-EAFLRFCKFEEKHKNIPRARAGYEKAIELLP 68
YI E + ++R R+++ER+L P A+ + +FE RARA +E AI P
Sbjct 449 YIEMELKLVNIDRCRKLYERFLEWSPENCYAWRNYAEFEISLAETERARAIFELAIS-QP 507
Query 69 EDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFGEHEG 128
+ E + + FE + E E+ +A+YE L R + ++ + F+ EH+
Sbjct 508 ALDMPELLWKTYIDFEISEGEFEKTRALYERLLDRTKHCK---VWISFAKFEASASEHKE 564
Query 129 GG 130
G
Sbjct 565 DG 566
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 37/141 (26%), Positives = 65/141 (46%), Gaps = 31/141 (21%)
Query 7 WLVYIHFEERCRELERARRIFERYLSNR-PSQEAFLRFCKFEEKHKNIPRARAGYEKAIE 65
W+ Y +EE + RAR ++ER L + ++++ +FE K+K + AR +++++
Sbjct 81 WVKYAKWEESQMDYARARSVWERALEGEYRNHTLWVKYAEFEMKNKFVNNARNVWDRSVT 140
Query 66 LLP--EDLLNEHFYMK----------------------------FAAFEERQQEKERAKA 95
LLP + L ++ YM+ F FE R E ERA++
Sbjct 141 LLPRVDQLWEKYIYMEEKLGNVTGARQIFERWMNWSPDQKAWLCFIKFELRYNEIERARS 200
Query 96 IYEAALQRVPRGQADELYSKY 116
IYE + P+ A Y+K+
Sbjct 201 IYERFVLCHPKVSAFIRYAKF 221
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 33/133 (24%), Positives = 64/133 (48%), Gaps = 16/133 (12%)
Query 1 NPSD-KGWLVYIHFEERCRELERARRIFERYLSNR-PSQE---------AFLRFCKFEE- 48
NP + W Y+ EE +R R I+ER ++N P+QE ++ + +EE
Sbjct 324 NPLNYDSWFDYVRLEESVGNKDRIREIYERAIANVPPAQEKRFWQRYIYLWINYALYEEI 383
Query 49 KHKNIPRARAGYEKAIELLPEDLLN-EHFYMKFAAFEERQQEKERAKAIYEAALQRVPRG 107
+ K++ R R Y + ++L+P + ++ A +E RQ A+ I A+ + P+
Sbjct 384 ETKDVERTRDVYRECLKLIPHTKFSFAKIWLLAAEYEIRQLNLTGARQILGNAIGKAPKV 443
Query 108 QADELYSKYVSFQ 120
+ ++ KY+ +
Sbjct 444 K---IFKKYIEME 453
Score = 32.0 bits (71), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 29/62 (46%), Gaps = 5/62 (8%)
Query 7 WLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKF-----EEKHKNIPRARAGYE 61
W YI FE E E+ R ++ER L + ++ F KF E K I AR ++
Sbjct 516 WKTYIDFEISEGEFEKTRALYERLLDRTKHCKVWISFAKFEASASEHKEDGIKSARVIFD 575
Query 62 KA 63
+A
Sbjct 576 RA 577
> dre:393920 crnkl1, MGC55327, zgc:55327; crooked neck pre-mRNA
splicing factor-like 1 (Drosophila); K12869 crooked neck
Length=753
Score = 110 bits (274), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 51/127 (40%), Positives = 85/127 (66%), Gaps = 0/127 (0%)
Query 2 PSDKGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGYE 61
P ++ W YI+FE R +E+++AR I+E ++ P + ++++ FEEKH + R R +E
Sbjct 179 PEEQAWHSYINFELRYKEVDKARSIYENFVMVHPEVKNWIKYAHFEEKHGYVARGRKVFE 238
Query 62 KAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQK 121
+A+E E+ ++E+ Y+ FA FEE+Q+E ER + IY+ AL R+P+ QA EL+ Y F+K
Sbjct 239 RAVEFFGEEQVSENLYVAFARFEEKQKEFERVRVIYKYALDRIPKQQAQELFKNYTVFEK 298
Query 122 QFGEHEG 128
+FG+ G
Sbjct 299 RFGDRRG 305
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 35/125 (28%), Positives = 62/125 (49%), Gaps = 3/125 (2%)
Query 5 KGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGYEKAI 64
K WL+Y FE R + L+ ARR + P + F + + E + + R R YEK +
Sbjct 412 KIWLLYGQFEIRQKNLQNARRGLGTAIGKCPKNKLFKGYIELELQLREFDRCRKLYEKYL 471
Query 65 ELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFG 124
E PE+ ++KFA E + +R++AI+E A+ + + L+ Y+ F+ +
Sbjct 472 EFSPENCTT---WIKFAELETILGDTDRSRAIFELAIGQPRLDMPEVLWKSYIDFEIEQE 528
Query 125 EHEGG 129
E++
Sbjct 529 EYDNT 533
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 32/121 (26%), Positives = 67/121 (55%), Gaps = 7/121 (5%)
Query 6 GWLVYIHFEERCRELERARRIFERYLS-NRPSQEAFLRFCKFEEKHKNIPRARAGYEKAI 64
W+ Y +EE +E++R+R I+ER L + + +L++ + E K++ + AR +++AI
Sbjct 82 NWIKYAQWEESLQEVQRSRSIYERALDVDHRNITLWLKYAEMEMKNRQVNHARNIWDRAI 141
Query 65 ELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFG 124
+LP +N+ F+ K+ EE + ++E ++ P QA + Y++F+ ++
Sbjct 142 TILPR--VNQ-FWYKYTYMEEMLGNIAGCRQVFERWMEWEPEEQA---WHSYINFELRYK 195
Query 125 E 125
E
Sbjct 196 E 196
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 34/120 (28%), Positives = 61/120 (50%), Gaps = 5/120 (4%)
Query 2 PSDKGWLVYIHFEERCRELERARRIFERYLSNRPSQ-EAFLRFCKFEEKHKNIPRARAGY 60
P +K + YI E + RE +R R+++E+YL P +++F + E + R+RA +
Sbjct 442 PKNKLFKGYIELELQLREFDRCRKLYEKYLEFSPENCTTWIKFAELETILGDTDRSRAIF 501
Query 61 EKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQ 120
E AI P + E + + FE Q+E + + +Y+ LQR Q +++ Y F+
Sbjct 502 ELAIG-QPRLDMPEVLWKSYIDFEIEQEEYDNTRGLYKRLLQRT---QHVKVWISYAQFE 557
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 23/96 (23%), Positives = 53/96 (55%), Gaps = 5/96 (5%)
Query 33 NRPSQEAFLRFCKFEEKHKNIPRARAGYEKAIELLPEDLLNEHFYMKFAAFEERQQEKER 92
NR ++++ ++EE + + R+R+ YE+A+++ D N ++K+A E + ++
Sbjct 76 NRTVISNWIKYAQWEESLQEVQRSRSIYERALDV---DHRNITLWLKYAEMEMKNRQVNH 132
Query 93 AKAIYEAALQRVPRGQADELYSKYVSFQKQFGEHEG 128
A+ I++ A+ +PR ++ + KY ++ G G
Sbjct 133 ARNIWDRAITILPR--VNQFWYKYTYMEEMLGNIAG 166
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 38/172 (22%), Positives = 68/172 (39%), Gaps = 45/172 (26%)
Query 1 NPSDKGWLVYIHFEERCRELERARRIFERYLS----NRPSQEAFLRFCKFEEKHKNIPRA 56
+P K W+ Y HFEE+ + R R++FER + + S+ ++ F +FEEK K R
Sbjct 211 HPEVKNWIKYAHFEEKHGYVARGRKVFERAVEFFGEEQVSENLYVAFARFEEKQKEFERV 270
Query 57 RAGYEKAIELLP------------------------EDLL--------------NEHFY- 77
R Y+ A++ +P ED++ N H Y
Sbjct 271 RVIYKYALDRIPKQQAQELFKNYTVFEKRFGDRRGIEDVIVSKRRFQYEEEVKANPHNYD 330
Query 78 --MKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFGEHE 127
+ E + + + +YE A+ +P Q + +Y+ + +E
Sbjct 331 AWFDYLRLVESDADADTVREVYERAIANIPPIQEKRHWRRYIYLWINYALYE 382
Score = 36.2 bits (82), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 30/138 (21%), Positives = 59/138 (42%), Gaps = 16/138 (11%)
Query 1 NPSD-KGWLVYIHFEERCRELERARRIFERYLSNRPSQE----------AFLRFCKFEEK 49
NP + W Y+ E + + R ++ER ++N P + ++ + +EE
Sbjct 325 NPHNYDAWFDYLRLVESDADADTVREVYERAIANIPPIQEKRHWRRYIYLWINYALYEEL 384
Query 50 HKNIP-RARAGYEKAIELLPEDLLN-EHFYMKFAAFEERQQEKERAKAIYEAALQRVPRG 107
P R R Y+ +EL+P ++ + FE RQ+ + A+ A+ + P+
Sbjct 385 EVKDPERTRQVYKACLELIPHKKFTFAKIWLLYGQFEIRQKNLQNARRGLGTAIGKCPKN 444
Query 108 QADELYSKYVSFQKQFGE 125
+ L+ Y+ + Q E
Sbjct 445 K---LFKGYIELELQLRE 459
> ath:AT3G51110 crooked neck protein, putative / cell cycle protein,
putative
Length=413
Score = 109 bits (273), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 55/129 (42%), Positives = 87/129 (67%), Gaps = 1/129 (0%)
Query 1 NPSDKGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGY 60
+P + WL +I FE R E+ER+R I+ER++ P +F+R+ KFE K+ + AR Y
Sbjct 170 SPDQQAWLCFIKFELRYNEIERSRSIYERFVLCHPKASSFIRYAKFEMKNSQVSLARIVY 229
Query 61 EKAIELLPE-DLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSF 119
E+AIE+L + + E ++ FA FEE +E ERA+ +Y+ AL +P+G+A++LY K+V+F
Sbjct 230 ERAIEMLKDVEEEAEMIFVAFAEFEELCKEVERARFLYKYALDHIPKGRAEDLYKKFVAF 289
Query 120 QKQFGEHEG 128
+KQ+G EG
Sbjct 290 EKQYGNKEG 298
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 26/96 (27%), Positives = 55/96 (57%), Gaps = 4/96 (4%)
Query 34 RPSQEAFLRFCKFEEKHKNIPRARAGYEKAIELLPEDLLNEHFYMKFAAFEERQQEKERA 93
+ + + ++R+ +EE K+ RAR+ +E+A+E E N ++K+A FE R + A
Sbjct 68 KTNSQVWVRYADWEESQKDHDRARSVWERALE--DESYRNHTLWLKYAEFEMRNKSVNHA 125
Query 94 KAIYEAALQRVPRGQADELYSKYVSFQKQFGEHEGG 129
+ +++ A++ +PR D+ + KY+ ++ G +G
Sbjct 126 RNVWDRAVKILPR--VDQFWYKYIHMEEILGNIDGA 159
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 32/128 (25%), Positives = 70/128 (54%), Gaps = 8/128 (6%)
Query 2 PSDKGWLVYIHFEERCRELERARRIFERYLSNRP--SQEAFLRFCKFEEKHKNIPRARAG 59
+ + W+ Y +EE ++ +RAR ++ER L + + +L++ +FE ++K++ AR
Sbjct 69 TNSQVWVRYADWEESQKDHDRARSVWERALEDESYRNHTLWLKYAEFEMRNKSVNHARNV 128
Query 60 YEKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSF 119
+++A+++LP + F+ K+ EE + A+ I+E + P QA + ++ F
Sbjct 129 WDRAVKILPR---VDQFWYKYIHMEEILGNIDGARKIFERWMDWSPDQQA---WLCFIKF 182
Query 120 QKQFGEHE 127
+ ++ E E
Sbjct 183 ELRYNEIE 190
> cel:M03F8.3 hypothetical protein; K12869 crooked neck
Length=747
Score = 100 bits (248), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 50/128 (39%), Positives = 84/128 (65%), Gaps = 1/128 (0%)
Query 2 PSDKGWLVYIHFEERCRELERARRIFERYL-SNRPSQEAFLRFCKFEEKHKNIPRARAGY 60
P ++ W YI+FE R +E++RAR +++R+L + + + ++++ KFEE++ I ARA Y
Sbjct 185 PPEQAWQTYINFELRYKEIDRARSVYQRFLHVHGINVQNWIKYAKFEERNGYIGNARAAY 244
Query 61 EKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQ 120
EKA+E E+ +NE + FA FEERQ+E ERA+ I++ L +P + +E++ Y +
Sbjct 245 EKAMEYFGEEDINETVLVAFALFEERQKEHERARGIFKYGLDNLPSNRTEEIFKHYTQHE 304
Query 121 KQFGEHEG 128
K+FGE G
Sbjct 305 KKFGERVG 312
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 37/119 (31%), Positives = 65/119 (54%), Gaps = 7/119 (5%)
Query 7 WLVYIHFEERCRELERARRIFERYLSNRP-SQEAFLRFCKFEEKHKNIPRARAGYEKAIE 65
WL Y E RC+++ AR +F+R ++ P + + +L++ EE +NIP AR +E+ IE
Sbjct 123 WLQYAEMEMRCKQINHARNVFDRAITIMPRAMQFWLKYSYMEEVIENIPGARQIFERWIE 182
Query 66 LLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFG 124
P E + + FE R +E +RA+++Y+ L G + + KY F+++ G
Sbjct 183 WEPP----EQAWQTYINFELRYKEIDRARSVYQRFLH--VHGINVQNWIKYAKFEERNG 235
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 35/123 (28%), Positives = 61/123 (49%), Gaps = 3/123 (2%)
Query 5 KGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGYEKAI 64
K W+++ HFE R +L AR+I + P + F + E + + R R YEK +
Sbjct 423 KVWIMFAHFEIRQLDLNAARKIMGVAIGKCPKDKLFRAYIDLELQLREFDRCRKLYEKFL 482
Query 65 ELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFG 124
E PE + ++KFA E + +R++A++ A+Q+ + L+ Y+ F+
Sbjct 483 ESSPE---SSQTWIKFAELETLLGDTDRSRAVFTIAVQQPALDMPELLWKAYIDFEIACE 539
Query 125 EHE 127
EHE
Sbjct 540 EHE 542
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 36/129 (27%), Positives = 67/129 (51%), Gaps = 5/129 (3%)
Query 2 PSDKGWLVYIHFEERCRELERARRIFERYLSNRP-SQEAFLRFCKFEEKHKNIPRARAGY 60
P DK + YI E + RE +R R+++E++L + P S + +++F + E + R+RA +
Sbjct 453 PKDKLFRAYIDLELQLREFDRCRKLYEKFLESSPESSQTWIKFAELETLLGDTDRSRAVF 512
Query 61 EKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQ 120
A++ P + E + + FE +E E+A+ +YE LQR + ++ F+
Sbjct 513 TIAVQ-QPALDMPELLWKAYIDFEIACEEHEKARDLYETLLQRTNHIK---VWISMAEFE 568
Query 121 KQFGEHEGG 129
+ G EG
Sbjct 569 QTIGNFEGA 577
Score = 54.7 bits (130), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 35/121 (28%), Positives = 67/121 (55%), Gaps = 7/121 (5%)
Query 6 GWLVYIHFEERCRELERARRIFERYLS-NRPSQEAFLRFCKFEEKHKNIPRARAGYEKAI 64
W+ Y +EE E++RAR +FER L + S +L++ + E + K I AR +++AI
Sbjct 88 NWIKYGKWEESIGEIQRARSVFERALDVDHRSISIWLQYAEMEMRCKQINHARNVFDRAI 147
Query 65 ELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFG 124
++P + F++K++ EE + A+ I+E ++ P QA + Y++F+ ++
Sbjct 148 TIMPRAM---QFWLKYSYMEEVIENIPGARQIFERWIEWEPPEQA---WQTYINFELRYK 201
Query 125 E 125
E
Sbjct 202 E 202
Score = 37.0 bits (84), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 29/57 (50%), Gaps = 0/57 (0%)
Query 7 WLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGYEKA 63
W YI FE C E E+AR ++E L + ++ +FE+ N AR +E+A
Sbjct 528 WKAYIDFEIACEEHEKARDLYETLLQRTNHIKVWISMAEFEQTIGNFEGARKAFERA 584
Score = 35.8 bits (81), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 25/98 (25%), Positives = 55/98 (56%), Gaps = 6/98 (6%)
Query 33 NRPSQEAFLRFCKFEEKHKNIPRARAGYEKAIELLPEDLLNEHFYMKFAAFEERQQEKER 92
NR ++++ K+EE I RAR+ +E+A+++ D + ++++A E R ++
Sbjct 82 NRMQLANWIKYGKWEESIGEIQRARSVFERALDV---DHRSISIWLQYAEMEMRCKQINH 138
Query 93 AKAIYEAALQRVPRGQADELYSKYVSFQKQFGEHEGGG 130
A+ +++ A+ +PR A + + KY S+ ++ E+ G
Sbjct 139 ARNVFDRAITIMPR--AMQFWLKY-SYMEEVIENIPGA 173
> ath:AT3G13210 crooked neck protein, putative / cell cycle protein,
putative
Length=657
Score = 90.9 bits (224), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 49/128 (38%), Positives = 78/128 (60%), Gaps = 10/128 (7%)
Query 1 NPSDKGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGY 60
+P + WL +I FE + E+E AR I+ER++ P A++R+ KFE KH + A +
Sbjct 155 SPDQQAWLCFIKFELKYNEIECARSIYERFVLCHPKVSAYIRYAKFEMKHGQVELAMKVF 214
Query 61 EKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQ 120
E+A + L +D E ++ FA FEE+ Y+ AL ++P+G+A+ LYSK+V+F+
Sbjct 215 ERAKKELADDEEAEILFVAFAEFEEQ----------YKFALDQIPKGRAENLYSKFVAFE 264
Query 121 KQFGEHEG 128
KQ G+ EG
Sbjct 265 KQNGDKEG 272
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 38/123 (30%), Positives = 60/123 (48%), Gaps = 3/123 (2%)
Query 5 KGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGYEKAI 64
K WL+ E R L AR+I + P + F ++ + E + +NI R R YE+ +
Sbjct 381 KIWLLAAQHEIRQLNLTGARQILGNAIGKAPKDKIFKKYIEIELQLRNIDRCRKLYERYL 440
Query 65 ELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFG 124
E P N + + K+A FE E ER +AI+E A+ + + L+ Y+ F+ G
Sbjct 441 EWSPG---NCYAWRKYAEFEMSLAETERTRAIFELAISQPALDMPELLWKTYIDFEISEG 497
Query 125 EHE 127
E E
Sbjct 498 ELE 500
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 35/104 (33%), Positives = 53/104 (50%), Gaps = 2/104 (1%)
Query 2 PSDKGWLVYIHFEERCRELERARRIFERYLSNRPSQ-EAFLRFCKFEEKHKNIPRARAGY 60
P DK + YI E + R ++R R+++ERYL P A+ ++ +FE R RA +
Sbjct 411 PKDKIFKKYIEIELQLRNIDRCRKLYERYLEWSPGNCYAWRKYAEFEMSLAETERTRAIF 470
Query 61 EKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRV 104
E AI P + E + + FE + E ER +A+YE L R
Sbjct 471 ELAIS-QPALDMPELLWKTYIDFEISEGELERTRALYERLLDRT 513
Score = 37.4 bits (85), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 24/94 (25%), Positives = 48/94 (51%), Gaps = 6/94 (6%)
Query 34 RPSQEAFLRFCKFEEKHKNIPRARAGYEKAIELLPEDLLNEHFYMKFAAFEERQQEKERA 93
R + + ++++ FE K+K++ AR +++A+ LLP + + KF EE+ A
Sbjct 88 RLNTQVWVKYADFEMKNKSVNEARNVWDRAVSLLPR---VDQLWYKFIHMEEKLGNIAGA 144
Query 94 KAIYEAALQRVPRGQADELYSKYVSFQKQFGEHE 127
+ I E + P QA + ++ F+ ++ E E
Sbjct 145 RQILERWIHCSPDQQA---WLCFIKFELKYNEIE 175
Score = 32.7 bits (73), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 17/41 (41%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 7 WLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFE 47
W YI FE ELER R ++ER L + ++ F KFE
Sbjct 486 WKTYIDFEISEGELERTRALYERLLDRTKHCKVWVDFAKFE 526
> sce:YLR117C CLF1, NTC77, SYF3; Clf1p; K12869 crooked neck
Length=687
Score = 67.8 bits (164), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 38/132 (28%), Positives = 65/132 (49%), Gaps = 10/132 (7%)
Query 2 PSDKGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGYE 61
P W ++ FE R + R I+ +Y+ P + +L++ +FE +H N R+ Y
Sbjct 164 PGVNAWNSFVDFEIRQKNWNGVREIYSKYVMAHPQMQTWLKWVRFENRHGNTEFTRSVYS 223
Query 62 KAIELLP--------EDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELY 113
AI+ + D+ FA +E QQE ER+ A+Y+ A+++ P Q L
Sbjct 224 LAIDTVANLQNLQIWSDMEVAKLVNSFAHWEAAQQEYERSSALYQIAIEKWPSNQL--LK 281
Query 114 SKYVSFQKQFGE 125
+ + F+KQFG+
Sbjct 282 AGLLDFEKQFGD 293
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 38/129 (29%), Positives = 58/129 (44%), Gaps = 7/129 (5%)
Query 5 KGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGYEKAI 64
K WL+Y F R ++ +AR+I + + P + F + + E K K R R YEK I
Sbjct 405 KIWLMYAKFLIRHDDVPKARKILGKAIGLCPKAKTFKGYIELEVKLKEFDRVRKIYEKFI 464
Query 65 ELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADE----LYSKYVSFQ 120
E P DL + ++ EE + +R + IY AL E L KY++F+
Sbjct 465 EFQPSDL---QIWSQYGELEENLGDWDRVRGIYTIALDENSDFLTKEAKIVLLQKYITFE 521
Query 121 KQFGEHEGG 129
+ E E
Sbjct 522 TESQEFEKA 530
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 51/100 (51%), Gaps = 4/100 (4%)
Query 2 PSDKGWLVYIHFEERCRELERARRIFERYLSNRPSQ-EAFLRFCKFEEKHKNIPRARAGY 60
P K + YI E + +E +R R+I+E+++ +PS + + ++ + EE + R R Y
Sbjct 435 PKAKTFKGYIELEVKLKEFDRVRKIYEKFIEFQPSDLQIWSQYGELEENLGDWDRVRGIY 494
Query 61 EKAIELLPEDLLNEH---FYMKFAAFEERQQEKERAKAIY 97
A++ + L E K+ FE QE E+A+ +Y
Sbjct 495 TIALDENSDFLTKEAKIVLLQKYITFETESQEFEKARKLY 534
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 35/122 (28%), Positives = 53/122 (43%), Gaps = 8/122 (6%)
Query 1 NPSDKG-WLVYIHFEERCRELERARRIFER-YLSNRPSQEAFLRFCKFEEKHKNIPRARA 58
N D G W+ Y FE ++ RAR IFER L + ++R+ E K K I AR
Sbjct 61 NRLDMGQWIRYAQFEIEQHDMRRARSIFERALLVDSSFIPLWIRYIDAELKVKCINHARN 120
Query 59 GYEKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVS 118
+AI LP + + K+ EE E +++Y P A ++ +V
Sbjct 121 LMNRAISTLPR---VDKLWYKYLIVEESLNNVEIVRSLYTKWCSLEPGVNA---WNSFVD 174
Query 119 FQ 120
F+
Sbjct 175 FE 176
> tgo:TGME49_105240 XPA-binding protein, putative ; K12867 pre-mRNA-splicing
factor SYF1
Length=966
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 39/126 (30%), Positives = 66/126 (52%), Gaps = 9/126 (7%)
Query 7 WLVYIH-FEER--CRELERARRIFERYLSNRPSQEA---FLRFCKFEEKHKNIPRARAGY 60
WL+Y+ F R +LERAR +F++ ++ P+Q A FL + K EE+ A Y
Sbjct 601 WLMYLTKFVSRYGSSKLERARELFQQATASVPAQHAKRFFLLYAKLEEEFGLAKHALTIY 660
Query 61 EKAIELLP-EDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSF 119
+ A + +P E+ L+ Y+ + A R + IYE A++ +P QA ++ +Y +
Sbjct 661 QAATKAVPQEEKLD--MYLIYIARTTELLGVARTRQIYEEAIENLPEKQARDMCLRYAAV 718
Query 120 QKQFGE 125
+K GE
Sbjct 719 EKGLGE 724
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 28/49 (57%), Gaps = 0/49 (0%)
Query 76 FYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFG 124
+M F F +RQ+ R + ++ ALQ + Q D+++ +Y+ F K+ G
Sbjct 137 IWMLFVDFLKRQKLLTRTRRAFDRALQSLAVTQHDQVWDRYIQFVKEAG 185
Score = 32.3 bits (72), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 18/68 (26%), Positives = 35/68 (51%), Gaps = 3/68 (4%)
Query 5 KGWLVYIHFEERCRELERARRIFERYLSNRPS---QEAFLRFCKFEEKHKNIPRARAGYE 61
K W++++ F +R + L R RR F+R L + + + R+ +F ++ + Y
Sbjct 136 KIWMLFVDFLKRQKLLTRTRRAFDRALQSLAVTQHDQVWDRYIQFVKEAGVVETTLRVYR 195
Query 62 KAIELLPE 69
+ + LLPE
Sbjct 196 RCLMLLPE 203
> ath:AT5G28740 transcription-coupled DNA repair protein-related;
K12867 pre-mRNA-splicing factor SYF1
Length=917
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 36/108 (33%), Positives = 58/108 (53%), Gaps = 4/108 (3%)
Query 12 HFEERCRELERARRIFERYLSNRPSQEAFL-RFCKFEEKHKNIPRARAGYEKAIELLPED 70
+FE+ + ER +IF +Y + +L +F K K K + RAR +E A+ + P D
Sbjct 564 YFEDAFKVYERGVKIF-KYPHVKDIWVTYLTKFVKRYGKTK-LERARELFEHAVSMAPSD 621
Query 71 LLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVS 118
+ Y+++A EE +RA +YE A ++VP GQ E+Y Y+S
Sbjct 622 AVRT-LYLQYAKLEEDYGLAKRAMKVYEEATKKVPEGQKLEMYEIYIS 668
Score = 36.6 bits (83), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 43/164 (26%), Positives = 68/164 (41%), Gaps = 44/164 (26%)
Query 7 WLVYIH-FEERC--RELERARRIFERYLSNRPSQEA---FLRFCKFEEKHK--------- 51
W+ Y+ F +R +LERAR +FE +S PS +L++ K EE +
Sbjct 588 WVTYLTKFVKRYGKTKLERARELFEHAVSMAPSDAVRTLYLQYAKLEEDYGLAKRAMKVY 647
Query 52 ---------------------------NIPRARAGYEKAIEL-LPEDLLNEHFYMKFAAF 83
+PR R YE+AIE LP + + +KFA
Sbjct 648 EEATKKVPEGQKLEMYEIYISRAAEIFGVPRTREIYEQAIESGLPHKDV-KIMCIKFAEL 706
Query 84 EERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFGEHE 127
E E +RA+A+Y+ + Q E ++K+ F+ Q G +
Sbjct 707 ERSLGEIDRARALYKYSSQFADPRSDPEFWNKWHEFEVQHGNED 750
> hsa:24148 PRPF6, ANT-1, C20orf14, Prp6, TOM, U5-102K, hPrp6;
PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae);
K12855 pre-mRNA-processing factor 6
Length=941
Score = 44.3 bits (103), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 32/109 (29%), Positives = 53/109 (48%), Gaps = 10/109 (9%)
Query 1 NP-SDKGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAG 59
NP S++ WL + E E ERARR+ + S+ P+ F++ K E NI A+
Sbjct 638 NPNSEEIWLAAVKLESENDEYERARRLLAKARSSAPTARVFMKSVKLEWVQDNIRAAQDL 697
Query 60 YEKAI---ELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVP 105
E+A+ E P+ +M EE+++ E+A+ Y L++ P
Sbjct 698 CEEALRHYEDFPK------LWMMKGQIEEQKEMMEKAREAYNQGLKKCP 740
> cpv:cgd7_3690 crooked neck protein HAT repeats ; K12869 crooked
neck
Length=736
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 22/72 (30%), Positives = 41/72 (56%), Gaps = 4/72 (5%)
Query 2 PSDKGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGYE 61
P ++ Y FEE C E++ AR + + + + P + F+ + KFE++HKN+ + E
Sbjct 201 PDPSIYIQYSKFEEECGEIKSARGVMKDLIISYPDESNFIEYIKFEQRHKNLFSS----E 256
Query 62 KAIELLPEDLLN 73
+ I +L E L++
Sbjct 257 QIINILSETLID 268
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 21/85 (24%), Positives = 42/85 (49%), Gaps = 4/85 (4%)
Query 4 DKGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGYEKA 63
D+ W+ YI E + R I+ +++ +P ++++ KFEE+ I AR +
Sbjct 170 DEFWIKYIQMELILKNYINVRHIYRKWIDWKPDPSIYIQYSKFEEECGEIKSARGVMKDL 229
Query 64 IELLPEDLLNEHFYMKFAAFEERQQ 88
I P+ E ++++ FE+R +
Sbjct 230 IISYPD----ESNFIEYIKFEQRHK 250
Score = 29.3 bits (64), Expect = 3.0, Method: Composition-based stats.
Identities = 24/100 (24%), Positives = 44/100 (44%), Gaps = 6/100 (6%)
Query 22 RARRIFERYLS-NRPSQEAFLRFCKFEEKHKNIPRARAGYEKAIELLPEDLLNEHFYMKF 80
R R+ FE + R +L + K+E NI +R+ +E+ I + E N + ++
Sbjct 86 RKRKEFEDSIRRKRWKISLYLSYAKWESLQNNIKNSRSIFERGILVNYE---NVRIWREY 142
Query 81 AAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQ 120
E A+ ++E +PR DE + KY+ +
Sbjct 143 IKLEITNGNINNARNLFERVTHLLPR--IDEFWIKYIQME 180
> hsa:653889 pre-mRNA-processing factor 6-like
Length=406
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 32/109 (29%), Positives = 53/109 (48%), Gaps = 10/109 (9%)
Query 1 NP-SDKGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAG 59
NP S++ WL + E E ERARR+ + S+ P+ F++ K E NI A+
Sbjct 103 NPNSEEIWLAAVKLESENDEYERARRLLAKARSSAPTARVFMKSVKLEWVQDNIRAAQDL 162
Query 60 YEKAI---ELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVP 105
E+A+ E P+ +M EE+++ E+A+ Y L++ P
Sbjct 163 CEEALRHYEDFPK------LWMMKGQIEEQKEMMEKAREAYNQGLKKCP 205
> dre:323855 c20orf14, fc12b02, prpf6, wu:fa05f07, wu:fc12b02,
zgc:65913; c20orf14 homolog (H. sapiens); K12855 pre-mRNA-processing
factor 6
Length=944
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 32/112 (28%), Positives = 51/112 (45%), Gaps = 8/112 (7%)
Query 1 NP-SDKGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAG 59
NP S++ WL + E E ERARR+ + S+ P+ F++ + E NI A
Sbjct 641 NPNSEEIWLAAVKLESENNEYERARRLLAKARSSAPTARVFMKSVRLEWVLGNI---EAA 697
Query 60 YEKAIELLP--EDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQA 109
+E E L ED +M EE+ + +RA+ Y L++ P +
Sbjct 698 HELCTEALKHYEDF--PKLWMMRGQIEEQSESIDRAREAYNQGLKKCPHSMS 747
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 33/114 (28%), Positives = 52/114 (45%), Gaps = 6/114 (5%)
Query 5 KGWLVYIHFEERCRELERARRIFERYLSNRP-SQEAFLRFCKFEEKHKNIPRARAGYEKA 63
K W++ EE+ ++RAR + + L P S +L + EEK + RARA EKA
Sbjct 713 KLWMMRGQIEEQSESIDRAREAYNQGLKKCPHSMSLWLLLSRLEEKVGQLTRARAILEKA 772
Query 64 IELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYV 117
P+ + +++ E R K A + ALQ P + L+S+ V
Sbjct 773 RLKNPQ---SPELWLESVRLEYRAGLKNIANTLMAKALQECP--NSGILWSEAV 821
Score = 29.6 bits (65), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 30/91 (32%), Positives = 41/91 (45%), Gaps = 10/91 (10%)
Query 7 WLVYIHFEERCRELERARRIFERY-LSNRPSQEAFLRFCKFEEKH--KNIPRARAGYEKA 63
WL+ EE+ +L RAR I E+ L N S E +L + E + KNI A KA
Sbjct 749 WLLLSRLEEKVGQLTRARAILEKARLKNPQSPELWLESVRLEYRAGLKNI--ANTLMAKA 806
Query 64 IELLPEDLLNEHFYMKFAAF-EERQQEKERA 93
++ P N A F E R Q K ++
Sbjct 807 LQECP----NSGILWSEAVFLEARPQRKTKS 833
> mmu:68879 Prpf6, 1190003A07Rik, 2610031L17Rik, ANT-1, MGC11655,
MGC36967, MGC38351, U5-102K; PRP6 pre-mRNA splicing factor
6 homolog (yeast); K12855 pre-mRNA-processing factor 6
Length=941
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 32/109 (29%), Positives = 52/109 (47%), Gaps = 10/109 (9%)
Query 1 NP-SDKGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAG 59
NP S++ WL + E E ERARR+ + S+ P+ F++ K E NI A+
Sbjct 638 NPNSEEIWLAAVKLESENNEYERARRLLAKARSSAPTARVFMKSVKLEWVLGNISAAQEL 697
Query 60 YEKAI---ELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVP 105
E+A+ E P+ +M EE+ + E+A+ Y L++ P
Sbjct 698 CEEALRHYEDFPK------LWMMKGQIEEQGELMEKAREAYNQGLKKCP 740
> xla:447198 prpf6, MGC80263; PRP6 pre-mRNA processing factor
6 homolog; K12855 pre-mRNA-processing factor 6
Length=948
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 31/107 (28%), Positives = 52/107 (48%), Gaps = 10/107 (9%)
Query 1 NP-SDKGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAG 59
NP S++ WL + E E ERARR+ + S+ P+ F++ K E NI A+
Sbjct 645 NPNSEEIWLAAVKLESENNEYERARRLLAKARSSAPTARVFMKSVKLEWVLGNIEAAQDL 704
Query 60 YEKAI---ELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQR 103
E+A+ E P+ +M EE+ ++ E+A+ Y L++
Sbjct 705 CEEALRHYEDFPK------LWMMKGQIEEQMEQTEKARDAYNQGLKK 745
Score = 31.2 bits (69), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 28/102 (27%), Positives = 44/102 (43%), Gaps = 4/102 (3%)
Query 5 KGWLVYIHFEERCRELERARRIFERYLSNRP-SQEAFLRFCKFEEKHKNIPRARAGYEKA 63
K W++ EE+ + E+AR + + L S +L + EEK + RARA EK+
Sbjct 717 KLWMMKGQIEEQMEQTEKARDAYNQGLKKCIHSTSLWLLLSRLEEKVGQLTRARAILEKS 776
Query 64 IELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVP 105
P+ +++ E R K A + ALQ P
Sbjct 777 RLKNPK---TPELWLESVRLEFRAGLKNIANTLMAKALQECP 815
> xla:100036801 xab2; XPA binding protein 2; K12867 pre-mRNA-splicing
factor SYF1
Length=838
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 34/114 (29%), Positives = 58/114 (50%), Gaps = 10/114 (8%)
Query 18 RELERARRIFERYLSNRP---SQEAFLRFCKFEEKHKNIPRARAGYEKAIELLPEDLLNE 74
++LERAR +FE+ L P ++ FL + K EE+H A A YE+A + + E
Sbjct 562 KKLERARDLFEQSLDGCPRKFAKNIFLLYAKLEEEHGLARHAMALYERATQAVET---GE 618
Query 75 HFYMKFAAFEERQQE---KERAKAIYEAALQRVPRGQADELYSKYVSFQKQFGE 125
+ M F + +R E + IYE A++ +P Q+ E+ ++ + + GE
Sbjct 619 QYEM-FNIYIKRAAEIYGVTHTRTIYERAIELLPDEQSREMCLRFADMECKLGE 671
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 25/125 (20%), Positives = 54/125 (43%), Gaps = 3/125 (2%)
Query 3 SDKGWLVYIHFEERCRELERARRIFERYLS-NRPSQEAFLRFCKFEEKHKNIPRARAGYE 61
S + W + EE + + +++R + + + + + + F E+H + YE
Sbjct 474 SLRVWSMLADLEESLGTFKSTKAVYDRIIDLHIATPQIVINYAMFLEEHNYFEESFKAYE 533
Query 62 KAIELLPEDLLNEHFYMKFAAFEERQQEK--ERAKAIYEAALQRVPRGQADELYSKYVSF 119
+ I L + + + + F R K ERA+ ++E +L PR A ++ Y
Sbjct 534 RGIALFRWPNVYDIWSTYLSKFIARYGGKKLERARDLFEQSLDGCPRKFAKNIFLLYAKL 593
Query 120 QKQFG 124
+++ G
Sbjct 594 EEEHG 598
> cel:C50F2.3 hypothetical protein; K12867 pre-mRNA-splicing factor
SYF1
Length=855
Score = 41.2 bits (95), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 29/121 (23%), Positives = 57/121 (47%), Gaps = 3/121 (2%)
Query 7 WLVYIHFEERCRELERARRIFERYLSNR-PSQEAFLRFCKFEEKHKNIPRARAGYEKAIE 65
W +Y +EE C +E R+++++ + R S + + + F E+++ A YEK I
Sbjct 491 WAMYADYEECCGTVESCRKVYDKMIELRVASPQMIMNYAMFLEENEYFELAFQAYEKGIA 550
Query 66 LLPEDLLNEHFYMKFAAFEERQQEK--ERAKAIYEAALQRVPRGQADELYSKYVSFQKQF 123
L + + + F +R K ERA+ ++E L+ P A ++ Y +++
Sbjct 551 LFKWPGVFDIWNTYLVKFIKRYGGKKLERARDLFEQCLENCPPTHAKYIFLLYAKLEEEH 610
Query 124 G 124
G
Sbjct 611 G 611
Score = 37.7 bits (86), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 35/122 (28%), Positives = 50/122 (40%), Gaps = 40/122 (32%)
Query 18 RELERARRIFERYLSNRPSQEA---FLRFCKFEEKHK----------------------- 51
++LERAR +FE+ L N P A FL + K EE+H
Sbjct 575 KKLERARDLFEQCLENCPPTHAKYIFLLYAKLEEEHGLARHALSIYNRACSGVDRADMHS 634
Query 52 -------------NIPRARAGYEKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYE 98
I + R +E+AI LPED + +++A E E +RA+AIY
Sbjct 635 MYNIYIKKVQEMYGIAQCRPIFERAISELPED-KSRAMSLRYAQLETTVGEIDRARAIYA 693
Query 99 AA 100
A
Sbjct 694 HA 695
Score = 30.8 bits (68), Expect = 0.96, Method: Compositional matrix adjust.
Identities = 21/61 (34%), Positives = 30/61 (49%), Gaps = 3/61 (4%)
Query 9 VYIHFEERCRELERARRIFERYLSNRP---SQEAFLRFCKFEEKHKNIPRARAGYEKAIE 65
+YI + + + R IFER +S P S+ LR+ + E I RARA Y A E
Sbjct 638 IYIKKVQEMYGIAQCRPIFERAISELPEDKSRAMSLRYAQLETTVGEIDRARAIYAHAAE 697
Query 66 L 66
+
Sbjct 698 I 698
> dre:794079 pdcd11, MGC162501, cb680, im:7148359, sb:cb680, wu:fc68c10;
programmed cell death 11; K14792 rRNA biogenesis
protein RRP5
Length=1816
Score = 41.2 bits (95), Expect = 8e-04, Method: Composition-based stats.
Identities = 24/121 (19%), Positives = 55/121 (45%), Gaps = 3/121 (2%)
Query 7 WLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGYEKAIEL 66
W+ ++ E + +++FER + + + K + I A + Y+ ++
Sbjct 1606 WVAMLNLENMYGTPDSLQKVFERAIQYCEPLLVYQQLADIYAKSEKIKEAESLYKSMVKR 1665
Query 67 LPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFGEH 126
+D + Y+ + F RQ++ + A A+ + ALQ + + +L +++ + QFG
Sbjct 1666 FRQD---KAVYLSYGTFLLRQRQSDAANALLQRALQSLSSKEHVDLIARFARLEFQFGNS 1722
Query 127 E 127
E
Sbjct 1723 E 1723
> mmu:67439 Xab2, 0610041O14Rik, AV025587; XPA binding protein
2; K12867 pre-mRNA-splicing factor SYF1
Length=855
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 30/125 (24%), Positives = 56/125 (44%), Gaps = 3/125 (2%)
Query 3 SDKGWLVYIHFEERCRELERARRIFERYLSNR-PSQEAFLRFCKFEEKHKNIPRARAGYE 61
S K W + EE + + +++R L R + + + + F E+HK + YE
Sbjct 483 SLKVWSMLADLEESLGTFQSTKAVYDRILDLRIATPQIVINYAMFLEEHKYFEESFKAYE 542
Query 62 KAIELLPEDLLNEHFYMKFAAFEER--QQEKERAKAIYEAALQRVPRGQADELYSKYVSF 119
+ I L +++ + F R ++ ERA+ ++E AL P A LY Y
Sbjct 543 RGISLFKWPNVSDIWSTYLTKFISRYGGRKLERARDLFEQALDGCPPKYAKTLYLLYAQL 602
Query 120 QKQFG 124
++++G
Sbjct 603 EEEWG 607
Score = 31.6 bits (70), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 30/124 (24%), Positives = 55/124 (44%), Gaps = 4/124 (3%)
Query 7 WLVYIHFEERCRELERARRIFERYL-SNRPSQ--EAFLRFCKFEEKHKNIPRARAGYEKA 63
+L+Y EE A +++R + P+Q + F + K + + R Y+KA
Sbjct 596 YLLYAQLEEEWGLARHAMAVYDRATRAVEPAQQYDMFNIYIKRAAEIYGVTHTRGIYQKA 655
Query 64 IELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQF 123
IE+L ++ E ++FA E + E +RA+AIY Q + + F+ +
Sbjct 656 IEVLSDEHARE-MCLRFADMECKLGEIDRARAIYSFCSQICDPRTTGAFWQTWKDFEVRH 714
Query 124 GEHE 127
G +
Sbjct 715 GNED 718
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 31/115 (26%), Positives = 54/115 (46%), Gaps = 12/115 (10%)
Query 18 RELERARRIFERYLSNRPSQEA---FLRFCKFEEKHKNIPRARAGYEKAIELLPE----D 70
R+LERAR +FE+ L P + A +L + + EE+ A A Y++A + D
Sbjct 571 RKLERARDLFEQALDGCPPKYAKTLYLLYAQLEEEWGLARHAMAVYDRATRAVEPAQQYD 630
Query 71 LLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFGE 125
+ N Y+K AA + IY+ A++ + A E+ ++ + + GE
Sbjct 631 MFN--IYIKRAA---EIYGVTHTRGIYQKAIEVLSDEHAREMCLRFADMECKLGE 680
> hsa:56949 XAB2, DKFZp762C1015, HCNP, HCRN, NTC90, SYF1; XPA
binding protein 2; K12867 pre-mRNA-splicing factor SYF1
Length=855
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 30/125 (24%), Positives = 56/125 (44%), Gaps = 3/125 (2%)
Query 3 SDKGWLVYIHFEERCRELERARRIFERYLSNR-PSQEAFLRFCKFEEKHKNIPRARAGYE 61
S K W + EE + + +++R L R + + + + F E+HK + YE
Sbjct 483 SLKVWSMLADLEESLGTFQSTKAVYDRILDLRIATPQIVINYAMFLEEHKYFEESFKAYE 542
Query 62 KAIELLPEDLLNEHFYMKFAAFEER--QQEKERAKAIYEAALQRVPRGQADELYSKYVSF 119
+ I L +++ + F R ++ ERA+ ++E AL P A LY Y
Sbjct 543 RGISLFKWPNVSDIWSTYLTKFIARYGGRKLERARDLFEQALDGCPPKYAKTLYLLYAQL 602
Query 120 QKQFG 124
++++G
Sbjct 603 EEEWG 607
Score = 32.3 bits (72), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 31/124 (25%), Positives = 55/124 (44%), Gaps = 4/124 (3%)
Query 7 WLVYIHFEERCRELERARRIFERYL-SNRPSQ--EAFLRFCKFEEKHKNIPRARAGYEKA 63
+L+Y EE A ++ER + P+Q + F + K + + R Y+KA
Sbjct 596 YLLYAQLEEEWGLARHAMAVYERATRAVEPAQQYDMFNIYIKRAAEIYGVTHTRGIYQKA 655
Query 64 IELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQF 123
IE+L ++ E ++FA E + E +RA+AIY Q + + F+ +
Sbjct 656 IEVLSDEHARE-MCLRFADMECKLGEIDRARAIYSFCSQICDPRTTGAFWQTWKDFEVRH 714
Query 124 GEHE 127
G +
Sbjct 715 GNED 718
Score = 31.6 bits (70), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 32/115 (27%), Positives = 54/115 (46%), Gaps = 12/115 (10%)
Query 18 RELERARRIFERYLSNRPSQEA---FLRFCKFEEKHKNIPRARAGYEKAIELLPE----D 70
R+LERAR +FE+ L P + A +L + + EE+ A A YE+A + D
Sbjct 571 RKLERARDLFEQALDGCPPKYAKTLYLLYAQLEEEWGLARHAMAVYERATRAVEPAQQYD 630
Query 71 LLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFGE 125
+ N Y+K AA + IY+ A++ + A E+ ++ + + GE
Sbjct 631 MFN--IYIKRAA---EIYGVTHTRGIYQKAIEVLSDEHAREMCLRFADMECKLGE 680
> mmu:328110 Prpf39, FLJ11128, MGC37077, Srcs1; PRP39 pre-mRNA
processing factor 39 homolog (yeast); K13217 pre-mRNA-processing
factor 39
Length=665
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 27/133 (20%), Positives = 64/133 (48%), Gaps = 12/133 (9%)
Query 1 NPSD-KGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFL-RFCKFEEKHKNIPRARA 58
NP D GW+ + + E+ L AR+ F+++ + P + ++ E++H NI ++
Sbjct 89 NPQDFTGWVYLLQYVEQENHLMAARKAFDKFFVHYPYCYGYWKKYADLEKRHDNIKQSDE 148
Query 59 GYEKAIELLPEDLLNEHFYMKFAAFEER------QQEKERAKAIYE-AALQRVPRGQADE 111
Y + ++ +P L+ ++ + F + Q+ + +E A L ++D+
Sbjct 149 VYRRGLQAIP---LSVDLWIHYINFLKETLDPGDQETNTTIRGTFEHAVLAAGTDFRSDK 205
Query 112 LYSKYVSFQKQFG 124
L+ Y++++ + G
Sbjct 206 LWEMYINWENEQG 218
Score = 32.7 bits (73), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 48/94 (51%), Gaps = 6/94 (6%)
Query 5 KGWLVYIHFEERCRELERARRIFER-YLSNRPSQEAFLRFCKFEEKHKNIPRARAGYEKA 63
K W Y+ FE ER +FER +S +E ++++ K+ E H +I R + +A
Sbjct 352 KNWKEYLEFEIENGTHERVVVLFERCVISCALYEEFWIKYAKYMENH-SIEGVRHVFSRA 410
Query 64 IEL-LPEDLLNEHFYMKFAAFEERQQEKERAKAI 96
+ LP+ + +M +AAFEE+Q A+ I
Sbjct 411 CTVHLPKKPM---AHMLWAAFEEQQGNINEARII 441
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 33/65 (50%), Gaps = 2/65 (3%)
Query 3 SDKGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEK-HKNIPRARAGYE 61
SDK W +YI++E L +++R L P+Q F +F+E N+PR E
Sbjct 203 SDKLWEMYINWENEQGNLREVTAVYDRILG-IPTQLYSHHFQRFKEHVQNNLPRDLLTGE 261
Query 62 KAIEL 66
+ I+L
Sbjct 262 QFIQL 266
> dre:387600 xab2, MGC198247, zgc:63498, zgc:63949; XPA binding
protein 2; K12867 pre-mRNA-splicing factor SYF1
Length=851
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 28/125 (22%), Positives = 55/125 (44%), Gaps = 3/125 (2%)
Query 3 SDKGWLVYIHFEERCRELERARRIFERYLSNR-PSQEAFLRFCKFEEKHKNIPRARAGYE 61
S K W + EE + + +++R + R + + + + F E+H + YE
Sbjct 478 SLKVWSMLADLEESLGTFQSTKAVYDRIIDLRIATPQIIINYAMFLEEHNYFEESFKAYE 537
Query 62 KAIELLPEDLLNEHFYMKFAAFEERQQEK--ERAKAIYEAALQRVPRGQADELYSKYVSF 119
+ I L +++ + F +R K ERA+ ++E AL P A +Y Y
Sbjct 538 RGIALFKWPNVHDIWNTYLTKFIDRYGGKKLERARDLFEQALDGCPAKYAKTIYLLYAKL 597
Query 120 QKQFG 124
++++G
Sbjct 598 EEEYG 602
Score = 37.0 bits (84), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 36/132 (27%), Positives = 57/132 (43%), Gaps = 41/132 (31%)
Query 18 RELERARRIFERYLSNRPSQEA---FLRFCKFEEKHK----------------------- 51
++LERAR +FE+ L P++ A +L + K EE++
Sbjct 566 KKLERARDLFEQALDGCPAKYAKTIYLLYAKLEEEYGLARHAMAVYERATAAVEAEERHQ 625
Query 52 -------------NIPRARAGYEKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYE 98
+ RA Y+KAIE+LP++ + ++FA E + E +RA+AIY
Sbjct 626 MFNIYIKRAAEIYGVTHTRAIYQKAIEVLPDEHARD-MCLRFADMESKLGEIDRARAIYS 684
Query 99 AALQRV-PRGQA 109
Q PR A
Sbjct 685 YCSQICDPRVTA 696
> xla:779090 pdcd11, alg-4, alg4, nfbp, rrp5; programmed cell
death 11; K14792 rRNA biogenesis protein RRP5
Length=1812
Score = 38.1 bits (87), Expect = 0.007, Method: Composition-based stats.
Identities = 39/157 (24%), Positives = 63/157 (40%), Gaps = 36/157 (22%)
Query 7 WLVYIHFEERCRELERARRIFERYL---SNRPSQE-----------------------AF 40
WL Y+ F E+E+AR + ER L S R QE AF
Sbjct 1563 WLQYMAFHLHATEIEKARVVAERALKTISFREEQEKLNVWVALLNLENMYGTEESLTKAF 1622
Query 41 LRFCKFEEKHKNIPRARAGYEKAIEL-LPEDLLN---------EHFYMKFAAFEERQQEK 90
R ++ E K + Y K+ + EDL N + ++KFA F +Q +
Sbjct 1623 ERAVQYNEPLKVFQQLADIYIKSEKFKQAEDLYNTMLKRFRQEKSVWIKFATFLLKQGQG 1682
Query 91 ERAKAIYEAALQRVPRGQADELYSKYVSFQKQFGEHE 127
+ + + AL+ +P ++ SK+ + Q G+ E
Sbjct 1683 DGTHKLLQRALKSLPEKDHVDVISKFAQLEFQLGDTE 1719
Score = 37.0 bits (84), Expect = 0.014, Method: Composition-based stats.
Identities = 24/96 (25%), Positives = 46/96 (47%), Gaps = 1/96 (1%)
Query 10 YIHFEERCRELERARRIFERYLSNRPSQ-EAFLRFCKFEEKHKNIPRARAGYEKAIELLP 68
+ E + + ERA+ +FE LS+ P + + + + KH + R +E+ I L
Sbjct 1708 FAQLEFQLGDTERAKALFESTLSSYPKRTDLWSVYIDMMVKHGSQKEVRDIFERVIHLSL 1767
Query 69 EDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRV 104
+ F+ ++ +E++ E +A+ E ALQ V
Sbjct 1768 AAKKIKFFFKRYLEYEKKHGSTESVQAVKEKALQYV 1803
Score = 35.4 bits (80), Expect = 0.045, Method: Composition-based stats.
Identities = 29/121 (23%), Positives = 57/121 (47%), Gaps = 10/121 (8%)
Query 9 VYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPRARAGYEKAIELLP 68
+YI E+ + + + +R+ R + +++F F K ++A++ LP
Sbjct 1641 IYIKSEKFKQAEDLYNTMLKRF---RQEKSVWIKFATFLLKQGQGDGTHKLLQRALKSLP 1697
Query 69 EDLLNEHFYM--KFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFGEH 126
E +H + KFA E + + ERAKA++E+ L P+ +L+S Y+ + G
Sbjct 1698 E---KDHVDVISKFAQLEFQLGDTERAKALFESTLSSYPK--RTDLWSVYIDMMVKHGSQ 1752
Query 127 E 127
+
Sbjct 1753 K 1753
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 19/63 (30%), Positives = 32/63 (50%), Gaps = 4/63 (6%)
Query 7 WLVYIHFEERCRELERARRIFERY----LSNRPSQEAFLRFCKFEEKHKNIPRARAGYEK 62
W VYI + + R IFER L+ + + F R+ ++E+KH + +A EK
Sbjct 1739 WSVYIDMMVKHGSQKEVRDIFERVIHLSLAAKKIKFFFKRYLEYEKKHGSTESVQAVKEK 1798
Query 63 AIE 65
A++
Sbjct 1799 ALQ 1801
> hsa:55015 PRPF39, FLJ11128, FLJ20666, FLJ45460, MGC149842, MGC149843;
PRP39 pre-mRNA processing factor 39 homolog (S. cerevisiae);
K13217 pre-mRNA-processing factor 39
Length=669
Score = 36.2 bits (82), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 28/130 (21%), Positives = 57/130 (43%), Gaps = 6/130 (4%)
Query 1 NPSD-KGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFL-RFCKFEEKHKNIPRARA 58
NP D GW+ + + E+ L AR+ F+R+ + P + ++ E++H NI +
Sbjct 91 NPQDFTGWVYLLQYVEQENHLMAARKAFDRFFIHYPYCYGYWKKYADLEKRHDNIKPSDE 150
Query 59 GYEKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIY----EAALQRVPRGQADELYS 114
Y + ++ +P + Y+ F + E I A L ++D L+
Sbjct 151 VYRRGLQAIPLSVDLWIHYINFLKETLDPGDPETNNTIRGTFEHAVLAAGTDFRSDRLWE 210
Query 115 KYVSFQKQFG 124
Y++++ + G
Sbjct 211 MYINWENEQG 220
Score = 33.5 bits (75), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 48/94 (51%), Gaps = 6/94 (6%)
Query 5 KGWLVYIHFEERCRELERARRIFER-YLSNRPSQEAFLRFCKFEEKHKNIPRARAGYEKA 63
K W Y+ FE ER +FER +S +E ++++ K+ E H +I R + +A
Sbjct 354 KNWKEYLEFEIENGTHERVVVLFERCVISCALYEEFWIKYAKYMENH-SIEGVRHVFSRA 412
Query 64 IEL-LPEDLLNEHFYMKFAAFEERQQEKERAKAI 96
+ LP+ + +M +AAFEE+Q A+ I
Sbjct 413 CTIHLPKKPM---VHMLWAAFEEQQGNINEARNI 443
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 33/65 (50%), Gaps = 2/65 (3%)
Query 3 SDKGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEK-HKNIPRARAGYE 61
SD+ W +YI++E L I++R L P+Q F +F+E N+PR E
Sbjct 205 SDRLWEMYINWENEQGNLREVTAIYDRILG-IPTQLYSHHFQRFKEHVQNNLPRDLLTGE 263
Query 62 KAIEL 66
+ I+L
Sbjct 264 QFIQL 268
> cel:Y59A8B.6 hypothetical protein; K12855 pre-mRNA-processing
factor 6
Length=968
Score = 36.2 bits (82), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 29/124 (23%), Positives = 56/124 (45%), Gaps = 6/124 (4%)
Query 2 PSDKGWLVYIHFEERCRELERARRIFERYLSNRPS-QEAFLRFCKFEEKHKNIPRARAGY 60
PS + W+ HFE +E A+R+ E + + +L + E+ ++ AR Y
Sbjct 700 PSARVWMKNAHFEWCLGNVEEAKRLCEECIQKYDDFHKIYLVLGQVLEQMNDVHGARLAY 759
Query 61 EKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQ 120
+ I P + ++ EE+ + +A+ E A R P+ D+L+ + V F+
Sbjct 760 TQGIRKCPGVI---PLWILLVRLEEKAGQIVKARVDLEKARLRNPKN--DDLWLESVRFE 814
Query 121 KQFG 124
++ G
Sbjct 815 QRVG 818
> hsa:22984 PDCD11, ALG-4, ALG4, KIAA0185, NFBP, RRP5; programmed
cell death 11; K14792 rRNA biogenesis protein RRP5
Length=1871
Score = 36.2 bits (82), Expect = 0.029, Method: Composition-based stats.
Identities = 29/125 (23%), Positives = 57/125 (45%), Gaps = 11/125 (8%)
Query 9 VYIHFEE---RCRELERARRIFERYLSN-RPSQEAFLRFCKFEEKHKNIPRARAGYEKAI 64
V++H + + + + A ++ R L R + ++++ F + + ++A+
Sbjct 1693 VFLHLADIYAKSEKFQEAGELYNRMLKRFRQEKAVWIKYGAFLLRRSQAAASHRVLQRAL 1752
Query 65 ELLPEDLLNEHF--YMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQ 122
E LP EH KFA E + + ERAKAI+E L P+ +++S Y+ +
Sbjct 1753 ECLPS---KEHVDVIAKFAQLEFQLGDAERAKAIFENTLSTYPK--RTDVWSVYIDMTIK 1807
Query 123 FGEHE 127
G +
Sbjct 1808 HGSQK 1812
Score = 31.6 bits (70), Expect = 0.66, Method: Composition-based stats.
Identities = 35/157 (22%), Positives = 64/157 (40%), Gaps = 36/157 (22%)
Query 7 WLVYIHFEERCRELERARRIFERYL---SNRPSQE-----------------------AF 40
WL Y+ F + E+E+AR + ER L S R QE F
Sbjct 1622 WLQYMAFHLQATEIEKARAVAERALKTISFREEQEKLNVWVALLNLENMYGSQESLTKVF 1681
Query 41 LRFCKFEEKHK------NIPRARAGYEKAIELLPEDL----LNEHFYMKFAAFEERQQEK 90
R ++ E K +I +++A EL L + ++K+ AF R+ +
Sbjct 1682 ERAVQYNEPLKVFLHLADIYAKSEKFQEAGELYNRMLKRFRQEKAVWIKYGAFLLRRSQA 1741
Query 91 ERAKAIYEAALQRVPRGQADELYSKYVSFQKQFGEHE 127
+ + + AL+ +P + ++ +K+ + Q G+ E
Sbjct 1742 AASHRVLQRALECLPSKEHVDVIAKFAQLEFQLGDAE 1778
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 27/123 (21%), Positives = 53/123 (43%), Gaps = 9/123 (7%)
Query 7 WLVYIHFEERCRELERARRIFERYLSNRPSQE---AFLRFCKFEEKHKNIPRARAGYEKA 63
W+ Y F R + + R+ +R L PS+E +F + E + + RA+A +E
Sbjct 1728 WIKYGAFLLRRSQAAASHRVLQRALECLPSKEHVDVIAKFAQLEFQLGDAERAKAIFENT 1787
Query 64 IELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAA--LQRVPRGQADELYSKYVSFQK 121
+ P+ Y+ Q++ + I+E L P+ + + +Y+ ++K
Sbjct 1788 LSTYPKRTDVWSVYIDMTIKHGSQKD---VRDIFERVIHLSLAPK-RMKFFFKRYLDYEK 1843
Query 122 QFG 124
Q G
Sbjct 1844 QHG 1846
> ath:AT4G03430 EMB2770 (EMBRYO DEFECTIVE 2770); RNA splicing
factor, transesterification mechanism; K12855 pre-mRNA-processing
factor 6
Length=1029
Score = 35.4 bits (80), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 29/104 (27%), Positives = 45/104 (43%), Gaps = 4/104 (3%)
Query 5 KGWLVYIHFEERCRELERARRIFERYLSNRP-SQEAFLRFCKFEEKHKNIPRARAGYEKA 63
K WL+ EER + LE+AR+ ++ L + P +L EEK + +ARA A
Sbjct 795 KLWLMLGQLEERFKHLEQARKAYDTGLKHCPHCIPLWLSLADLEEKVNGLNKARAILTTA 854
Query 64 IELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRG 107
+ P ++ E R K A+ + ALQ P+
Sbjct 855 RKKNPG---GAELWLAAIRAELRHDNKREAEHLMSKALQDCPKS 895
Score = 27.7 bits (60), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 34/139 (24%), Positives = 53/139 (38%), Gaps = 37/139 (26%)
Query 3 SDKGWLVYIHFEERCRELERARR-------------------IFERYLSN-----RPSQE 38
S++ WL E +E ERAR I ER L N R E
Sbjct 726 SEEIWLAAFKLEFENKEPERARMLLAKARERGGTERVWMKSAIVERELGNVEEERRLLNE 785
Query 39 AFLRFCKF----------EEKHKNIPRARAGYEKAIELLPEDLLNEHFYMKFAAFEERQQ 88
+F F EE+ K++ +AR Y+ ++ P + ++ A EE+
Sbjct 786 GLKQFPTFFKLWLMLGQLEERFKHLEQARKAYDTGLKHCPHCI---PLWLSLADLEEKVN 842
Query 89 EKERAKAIYEAALQRVPRG 107
+A+AI A ++ P G
Sbjct 843 GLNKARAILTTARKKNPGG 861
> mmu:381510 Dpy19l4, Gm1023, Narg3; dpy-19-like 4 (C. elegans)
Length=722
Score = 34.7 bits (78), Expect = 0.075, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 24/40 (60%), Gaps = 0/40 (0%)
Query 82 AFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQK 121
+ E RQ++K+R EA +++ R A E KYVSFQ+
Sbjct 8 SVEPRQRKKQRTSGSQEAKAEKIRRTPAPERAPKYVSFQR 47
> cpv:cgd5_920 Pre-mRNA splicing factor Pro1/Prp6. HAT repeat
protein ; K12855 pre-mRNA-processing factor 6
Length=923
Score = 34.3 bits (77), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 38/79 (48%), Gaps = 5/79 (6%)
Query 49 KHKNIPRARAGYEKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQ 108
K NI ARA +E + ++ E+F++K+A FEE+ E+ + + +L+ P Q
Sbjct 515 KSNNIISARAMFESSADMFKS---KEYFWIKWANFEEKYGNFEKVDHVLQKSLKNCPDKQ 571
Query 109 ADELYSKYVSFQKQFGEHE 127
L+ K Q G E
Sbjct 572 I--LWLKAAQNQSANGNAE 588
Score = 33.1 bits (74), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 27/103 (26%), Positives = 44/103 (42%), Gaps = 3/103 (2%)
Query 7 WLVYIHFEERCRELERARRIFERYLSNRPSQEA-FLRFCKFEEKHKNIPRARAGYEKAIE 65
W+ + +FEE+ E+ + ++ L N P ++ +L+ + + + N AR K
Sbjct 540 WIKWANFEEKYGNFEKVDHVLQKSLKNCPDKQILWLKAAQNQSANGNAEIARLILSKGYS 599
Query 66 LLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQ 108
D E ++ A E Q E ERAK I E P Q
Sbjct 600 SSLND--KEEIVLEAARLELSQGEIERAKIILERERTNSPSVQ 640
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 19 ELERARRIFERYLSNRPSQEAFLRFCKFEEKHKN 52
E+ERA+ I ER +N PS + ++ K E KN
Sbjct 621 EIERAKIILERERTNSPSVQIWVESIKLENDQKN 654
> sce:YDR416W SYF1, NTC90; Member of the NineTeen Complex (NTC)
that contains Prp19p and stabilizes U6 snRNA in catalytic
forms of the spliceosome containing U2, U5, and U6 snRNAs; null
mutant has splicing defect and arrests in G2/M; homologs
in human and C. elegans; K12867 pre-mRNA-splicing factor SYF1
Length=859
Score = 33.5 bits (75), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 21/72 (29%), Positives = 36/72 (50%), Gaps = 1/72 (1%)
Query 56 ARAGYEKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSK 115
R Y++ I++LP E F +KF+ FE E RA+ I + +P + EL+
Sbjct 725 TRELYQECIQILPNSKAVE-FVIKFSDFESSIGETIRAREILAYGAKLLPPSRNTELWDS 783
Query 116 YVSFQKQFGEHE 127
+ F+ + G+ E
Sbjct 784 FEIFELKHGDKE 795
> mmu:18572 Pdcd11, 1110021I22Rik, ALG-4, Pdcd7, mKIAA0185; programmed
cell death 11; K14792 rRNA biogenesis protein RRP5
Length=1862
Score = 33.1 bits (74), Expect = 0.22, Method: Composition-based stats.
Identities = 29/124 (23%), Positives = 56/124 (45%), Gaps = 11/124 (8%)
Query 9 VYIHFEE---RCRELERARRIFERYLSN-RPSQEAFLRFCKFEEKHKNIPRARAGYEKAI 64
V++H + + + + A ++ R L R + ++++ F + ++A+
Sbjct 1684 VFLHLADIYTKSEKYKEAGELYNRMLKRFRQEKAVWIKYGAFVLGRSQAGASHRVLQRAL 1743
Query 65 ELLPEDLLNEHF--YMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQ 122
E LP EH +KFA E + + ERAKAI+E L P+ +++S Y+ +
Sbjct 1744 ECLPA---KEHVDVIVKFAQLEFQLGDVERAKAIFENTLSTYPK--RTDVWSVYIDMTIK 1798
Query 123 FGEH 126
G
Sbjct 1799 HGSQ 1802
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 29/121 (23%), Positives = 55/121 (45%), Gaps = 16/121 (13%)
Query 7 WLVYIHFEERCRELERARRIFERYL---SNRPSQE---AFLRFCKFEEKHKNIPRARAGY 60
WL Y+ F + E+E+AR + ER L S R QE ++ E + + +
Sbjct 1613 WLQYMAFHLQATEIEKARAVAERALKTISFREEQEKLNVWVALLNLENMYGSQESLTKVF 1672
Query 61 EKAIELLPEDLLNE--HFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVS 118
E+A++ NE ++ A + ++ + A +Y L+R + +A ++ KY +
Sbjct 1673 ERAVQ------YNEPLKVFLHLADIYTKSEKYKEAGELYNRMLKRFRQEKA--VWIKYGA 1724
Query 119 F 119
F
Sbjct 1725 F 1725
> dre:368864 prpf39, fa10d07, si:dz261o22.3, wu:fa10d07; PRP39
pre-mRNA processing factor 39 homolog (yeast); K13217 pre-mRNA-processing
factor 39
Length=752
Score = 33.1 bits (74), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 31/103 (30%), Positives = 51/103 (49%), Gaps = 6/103 (5%)
Query 5 KGWLVYIHFEERCRELERARRIFERYLSNRPSQEAF-LRFCKFEEKHKNIPRARAGYEKA 63
W Y+ FE ER +FER L E F +++ K+ E + + R Y+KA
Sbjct 429 NNWREYLDFELENGTPERVVVLFERCLIACALYEEFWIKYAKYLESY-STEAVRHIYKKA 487
Query 64 IEL-LPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVP 105
+ LP+ + ++ +AAFEE+Q + A++I +A VP
Sbjct 488 CTVHLPK---KPNVHLLWAAFEEQQGSIDEARSILKAVEVSVP 527
Score = 32.7 bits (73), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 33/127 (25%), Positives = 58/127 (45%), Gaps = 15/127 (11%)
Query 7 WLVYIHFEERCRELERARRIFERYLSNRP-SQEAFLRFCKFEEKHKNIP------RARAG 59
W Y E + ++ A ++ R L P S + +L + F ++++ R RA
Sbjct 203 WKKYADIERKHGYIQMADEVYRRGLQAIPLSVDLWLHYITFLRENQDTSDGEAESRIRAS 262
Query 60 YEKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSF 119
YE A+ D ++ + + A+E Q + AIY+ L +P +LYS++ F
Sbjct 263 YEHAVLACGTDFRSDRLWEAYIAWETEQGKLANVTAIYDRLL-CIPT----QLYSQH--F 315
Query 120 QKQFGEH 126
QK F +H
Sbjct 316 QK-FKDH 321
> pfa:PFL1735c RNA-processing protein, putative; K12867 pre-mRNA-splicing
factor SYF1
Length=1031
Score = 32.7 bits (73), Expect = 0.27, Method: Composition-based stats.
Identities = 23/92 (25%), Positives = 44/92 (47%), Gaps = 1/92 (1%)
Query 36 SQEAFLRFCKFEEKHKNIPRARAGYEKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKA 95
++ FL + FE + I + + Y++AI L E+ FY F + R ++A+
Sbjct 840 AKHIFLMYANFESNYGFIKKELSIYKEAIPFL-EEPDKIKFYKIFISKVSRAYGIQKARE 898
Query 96 IYEAALQRVPRGQADELYSKYVSFQKQFGEHE 127
+E A+Q + A +L Y+ + + E+E
Sbjct 899 AFEEAIQTLSDDSARQLCMIYIDMEYKLNEYE 930
> ath:AT1G08440 hypothetical protein
Length=501
Score = 32.3 bits (72), Expect = 0.34, Method: Composition-based stats.
Identities = 14/36 (38%), Positives = 24/36 (66%), Gaps = 3/36 (8%)
Query 18 RELERARRIFERY---LSNRPSQEAFLRFCKFEEKH 50
+E+E+ RR ERY L+++ ++EA F K+E +H
Sbjct 221 KEVEKRRRNLERYKSVLNSKSNEEALANFAKWEPRH 256
> cel:F25B4.5 hypothetical protein; K13217 pre-mRNA-processing
factor 39
Length=710
Score = 32.3 bits (72), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 29/124 (23%), Positives = 59/124 (47%), Gaps = 15/124 (12%)
Query 6 GWLVYIHFEERCRELERARRIFERYLSNRPSQEAFL-RFCKFEEKHKNIPRARAGYEKAI 64
W+ + ++ +++ AR + +LS P+ F ++ ++E+K NI A+A +EK I
Sbjct 101 NWVNILAKVDQSDDVDFAREKYRSFLSRYPNCYGFWQKYAEYEKKMGNIAEAKAVWEKGI 160
Query 65 ELLPEDLLNEHFYMKFAA-------FEERQQEKERAKAIYEAALQRVPRGQADELYSKYV 117
+P L+ ++ + A F A+AI A L+ Q+D L+ + +
Sbjct 161 ISIP---LSIDLWLGYTADVKNIKNFPPESLRDLYARAIEIAGLEY----QSDRLWLEAI 213
Query 118 SFQK 121
F++
Sbjct 214 GFER 217
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 27/133 (20%), Positives = 55/133 (41%), Gaps = 13/133 (9%)
Query 7 WLVYIHFEERCRELERARRIFERYLSNRP-SQEAFLRFCKFEEKHKNIP--RARAGYEKA 63
W Y +E++ + A+ ++E+ + + P S + +L + + KN P R Y +A
Sbjct 136 WQKYAEYEKKMGNIAEAKAVWEKGIISIPLSIDLWLGYTADVKNIKNFPPESLRDLYARA 195
Query 64 IELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRV----------PRGQADELY 113
IE+ + ++ +++ FE E K A+ +R+ P A +
Sbjct 196 IEIAGLEYQSDRLWLEAIGFERAVYMDELCKGNTNASCKRIGVLFDKLLSTPTFHAPSHF 255
Query 114 SKYVSFQKQFGEH 126
+YV + H
Sbjct 256 DRYVQYLNTIEPH 268
> cel:C16A3.3 let-716; LEThal family member (let-716); K14792
rRNA biogenesis protein RRP5
Length=1743
Score = 32.0 bits (71), Expect = 0.44, Method: Composition-based stats.
Identities = 28/135 (20%), Positives = 56/135 (41%), Gaps = 8/135 (5%)
Query 1 NPSD-----KGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKHKNIPR 55
NP++ K W Y++ E + +++FER N + K +K +
Sbjct 1517 NPTESDELLKIWTAYLNMEVAYGDAATVQKVFERACKNANAYTVHKTLSKIYQKFEKNAE 1576
Query 56 ARAGYEKAIELLPEDLLNEHFYMKFAAFEERQQEKERAKAIYEAALQRVPRGQAD-ELYS 114
A E+ ++ + L + A Q +++ A+ + AL+ P+ Q +L S
Sbjct 1577 ATQILEQMVKKFRANQL--EVWTLLAEHLMTQNDQKAARELLPRALKSAPKAQQHVQLIS 1634
Query 115 KYVSFQKQFGEHEGG 129
K+ + + G+ E G
Sbjct 1635 KFAQLEFKHGDAERG 1649
> xla:734636 prpf39.1, MGC115228, prpf39; PRP39 pre-mRNA processing
factor 39 homolog, gene 1; K13217 pre-mRNA-processing
factor 39
Length=641
Score = 32.0 bits (71), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 24/65 (36%), Positives = 33/65 (50%), Gaps = 2/65 (3%)
Query 3 SDKGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEK-HKNIPRARAGYE 61
SDK W +YI++E L I+ R L P+Q L F +F+E ++PR E
Sbjct 180 SDKLWEMYINWETEQGNLSGVTSIYSRLLG-IPTQFYSLHFQRFKEHIQGHLPREFLTSE 238
Query 62 KAIEL 66
K IEL
Sbjct 239 KFIEL 243
Score = 27.7 bits (60), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 26/95 (27%), Positives = 48/95 (50%), Gaps = 8/95 (8%)
Query 5 KGWLVYIHFEERCRELERARRIFER-YLSNRPSQEAFLRFCKFEEKHKNIPRARAGYEKA 63
W Y+ FE ER +FER ++ +E ++++ K+ E H ++ R Y +A
Sbjct 325 NNWKEYLEFELENGSNERIVILFERCVIACACYEEFWIKYAKYMENH-SVEGVRHVYNRA 383
Query 64 --IELLPEDLLNEHFYMKFAAFEERQQEKERAKAI 96
+ L + ++ ++ +AAFEE+Q E A+ I
Sbjct 384 CHVHLAKKPMV----HLLWAAFEEQQGNLEEARRI 414
> bbo:BBOV_III009870 17.m07855; hypothetical protein
Length=950
Score = 32.0 bits (71), Expect = 0.55, Method: Composition-based stats.
Identities = 17/66 (25%), Positives = 33/66 (50%), Gaps = 4/66 (6%)
Query 4 DKGWLVYIHFEERCRELERARRIFERYLSNRPSQEAFLRFCKFEEKH----KNIPRARAG 59
D+ WL +I + R + ++E + N PS E ++ + +FE + NI R R
Sbjct 69 DRFWLSWIEDKRRDSSDDALISMYECAVDNEPSAELWIAYTRFERRRASADANIGRIRTL 128
Query 60 YEKAIE 65
+E+ ++
Sbjct 129 FERGLK 134
> ath:AT5G16840 BPA1; BPA1 (BINDINGPARTNEROFACD11 1); nucleic
acid binding / nucleotide binding / oxidoreductase
Length=260
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 26/89 (29%), Positives = 41/89 (46%), Gaps = 12/89 (13%)
Query 37 QEAFLRFCKFEEKHKNIPRARAGY---EKAIEL-----LPEDLLNEHFYMKFAAFEERQQ 88
++A + F+EKH A AG ++ I L L+NE K A ++ Q
Sbjct 117 KDAVGKAKAFDEKHGFTSTATAGVASLDQKIGLSQKLTAGTSLVNE----KIKAVDQNFQ 172
Query 89 EKERAKAIYEAALQRVPRGQADELYSKYV 117
ER K++Y AA Q V + + ++YV
Sbjct 173 VTERTKSVYAAAEQTVSSAGSAVMKNRYV 201
> dre:503773 MGC158780, id:ibd5127, wu:fa56d01, zgc:158780; zgc:113159
Length=598
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 17/48 (35%), Positives = 26/48 (54%), Gaps = 0/48 (0%)
Query 79 KFAAFEERQQEKERAKAIYEAALQRVPRGQADELYSKYVSFQKQFGEH 126
+ EE + ++ KA YEA L R+ GQA + S +S Q+Q E+
Sbjct 110 ELTPIEEASRLNQKLKANYEARLARLTPGQATQKTSLTLSLQRQMMEN 157
> cpv:cgd7_970 Syf1p. protein with 8 HAT domains ; K12867 pre-mRNA-splicing
factor SYF1
Length=1020
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 19/71 (26%), Positives = 36/71 (50%), Gaps = 5/71 (7%)
Query 10 YIHFEERCRELERARRIFERYLSNRPSQEAF----LRFCKFEEKHKNIPRARAGYEKAIE 65
+I +CR++ R+IF+ + + + + LR+ FE + R R+ + A +
Sbjct 806 WIKLTLKCRDISYTRKIFDMAIDDIKASDIIIRLSLRYINFELNMGEVNRVRSIFIFAGD 865
Query 66 LLPE-DLLNEH 75
L+P L+NEH
Sbjct 866 LIPNIYLMNEH 876
> hsa:720 C4A, C4, C4A2, C4A3, C4A4, C4A6, C4S, CO4, CPAMD2, MGC164979,
RG; complement component 4A (Rodgers blood group);
K03989 complement component 4
Length=1744
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 28/41 (68%), Gaps = 3/41 (7%)
Query 37 QEAFLRFCKFEE--KHKNIPRARAGYEKAIELL-PEDLLNE 74
+E FL C+F E + K+ + +AG ++A+E+L EDL++E
Sbjct 729 REPFLSCCQFAESLRKKSRDKGQAGLQRALEILQEEDLIDE 769
> hsa:100507685 complement C4-B-like
Length=1744
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 28/41 (68%), Gaps = 3/41 (7%)
Query 37 QEAFLRFCKFEE--KHKNIPRARAGYEKAIELL-PEDLLNE 74
+E FL C+F E + K+ + +AG ++A+E+L EDL++E
Sbjct 729 REPFLSCCQFAESLRKKSRDKGQAGLQRALEILQEEDLIDE 769
> hsa:721 C4B, C4B1, C4B12, C4B2, C4B3, C4B5, C4F, CH, CO4, CPAMD3,
FLJ60561, MGC164979; complement component 4B (Chido blood
group); K03989 complement component 4
Length=1744
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 28/41 (68%), Gaps = 3/41 (7%)
Query 37 QEAFLRFCKFEE--KHKNIPRARAGYEKAIELL-PEDLLNE 74
+E FL C+F E + K+ + +AG ++A+E+L EDL++E
Sbjct 729 REPFLSCCQFAESLRKKSRDKGQAGLQRALEILQEEDLIDE 769
Lambda K H
0.320 0.137 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2039374804
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40