bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2553_orf1
Length=128
Score E
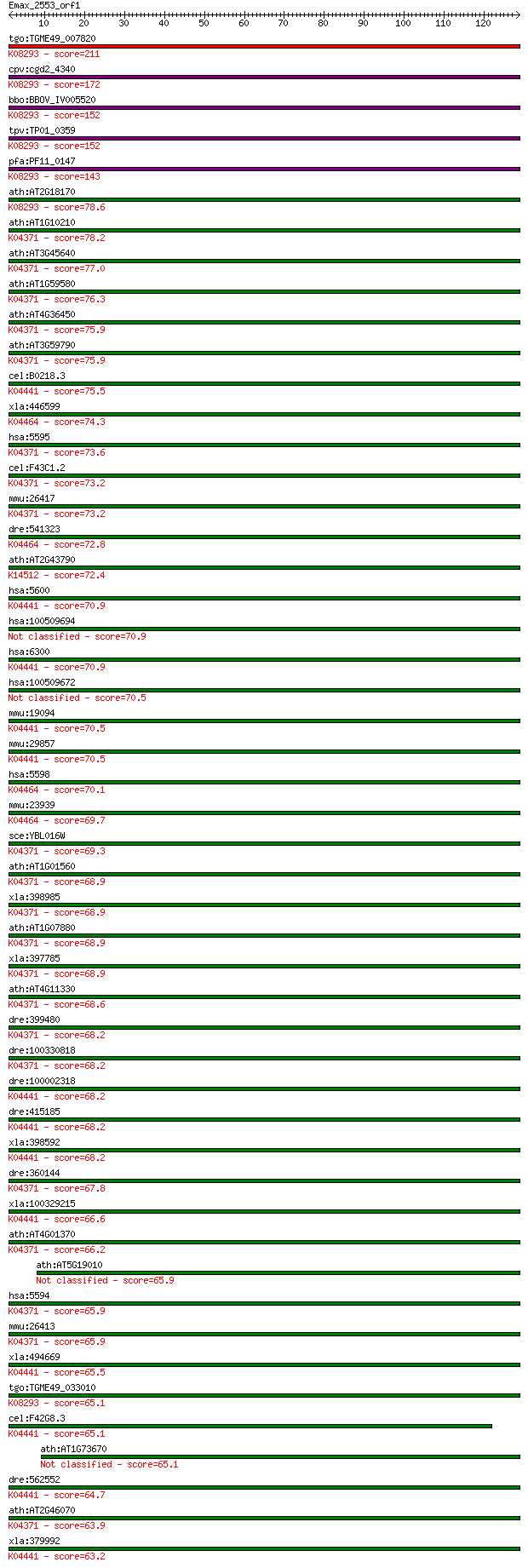
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_007820 mitogen-activated protein kinase, putative (... 211 7e-55
cpv:cgd2_4340 mitogen-activated protein kinase 2 ; K08293 mito... 172 2e-43
bbo:BBOV_IV005520 23.m06268; mitogen-activated protein kinase;... 152 3e-37
tpv:TP01_0359 mitogen-activated protein kinase 2; K08293 mitog... 152 3e-37
pfa:PF11_0147 map-2; mitogen-activated protein kinase 2; K0829... 143 1e-34
ath:AT2G18170 ATMPK7; ATMPK7 (ARABIDOPSIS THALIANA MAP KINASE ... 78.6 4e-15
ath:AT1G10210 ATMPK1; ATMPK1 (MITOGEN-ACTIVATED PROTEIN KINASE... 78.2 6e-15
ath:AT3G45640 ATMPK3; ATMPK3 (ARABIDOPSIS THALIANA MITOGEN-ACT... 77.0 1e-14
ath:AT1G59580 ATMPK2; ATMPK2 (ARABIDOPSIS THALIANA MITOGEN-ACT... 76.3 3e-14
ath:AT4G36450 ATMPK14 (Mitogen-activated protein kinase 14); M... 75.9 3e-14
ath:AT3G59790 ATMPK10; MAP kinase/ kinase; K04371 extracellula... 75.9 3e-14
cel:B0218.3 pmk-1; P38 Map Kinase family member (pmk-1); K0444... 75.5 4e-14
xla:446599 mapk7, MGC81757, bmk1, erk4, prkm7; mitogen-activat... 74.3 1e-13
hsa:5595 MAPK3, ERK1, HS44KDAP, HUMKER1A, MGC20180, P44ERK1, P... 73.6 1e-13
cel:F43C1.2 mpk-1; MAP Kinase family member (mpk-1); K04371 ex... 73.2 2e-13
mmu:26417 Mapk3, Erk-1, Erk1, Ert2, Esrk1, Mnk1, Mtap2k, Prkm3... 73.2 2e-13
dre:541323 mapk7, bmk1, erk5, wu:fb73b02, zgc:113111; mitogen-... 72.8 3e-13
ath:AT2G43790 ATMPK6; ATMPK6 (ARABIDOPSIS THALIANA MAP KINASE ... 72.4 3e-13
hsa:5600 MAPK11, P38B, P38BETA2, PRKM11, SAPK2, SAPK2B, p38-2,... 70.9 9e-13
hsa:100509694 mitogen-activated protein kinase 12-like 70.9 1e-12
hsa:6300 MAPK12, ERK3, ERK6, P38GAMMA, PRKM12, SAPK-3, SAPK3; ... 70.9 1e-12
hsa:100509672 mitogen-activated protein kinase 12-like 70.5 1e-12
mmu:19094 Mapk11, DKFZp586C1322, P38b, Prkm11, Sapk2, Sapk2b, ... 70.5 1e-12
mmu:29857 Mapk12, AW123708, Erk6, P38gamma, Prkm12, Sapk3; mit... 70.5 1e-12
hsa:5598 MAPK7, BMK1, ERK4, ERK5, PRKM7; mitogen-activated pro... 70.1 1e-12
mmu:23939 Mapk7, BMK1, ERK5, Erk5-T, PRKM7; mitogen-activated ... 69.7 2e-12
sce:YBL016W FUS3, DAC2; Fus3p (EC:2.7.11.24); K04371 extracell... 69.3 2e-12
ath:AT1G01560 ATMPK11; MAP kinase/ kinase; K04371 extracellula... 68.9 3e-12
xla:398985 mapk1-a, MGC68934, erk, erk2, mapk, mapk1, mapk1a, ... 68.9 4e-12
ath:AT1G07880 ATMPK13; MAP kinase/ kinase; K04371 extracellula... 68.9 4e-12
xla:397785 mapk1-b, Xp42, erk, erk2, mapk, mapk1b, mpk1; mitog... 68.9 4e-12
ath:AT4G11330 ATMPK5; ATMPK5 (MAP KINASE 5); MAP kinase/ kinas... 68.6 5e-12
dre:399480 mapk3, ERK1, fi06b09, wu:fi06b09, zERK1; mitogen-ac... 68.2 6e-12
dre:100330818 mitogen-activated protein kinase 3-like; K04371 ... 68.2 6e-12
dre:100002318 mapk13, im:7136778; mitogen-activated protein ki... 68.2 6e-12
dre:415185 zgc:86905; K04441 p38 MAP kinase [EC:2.7.11.24] 68.2 6e-12
xla:398592 mapk12, Xp38gamma, sapk3; mitogen-activated protein... 68.2 6e-12
dre:360144 mapk1, zERK2; mitogen-activated protein kinase 1 (E... 67.8 8e-12
xla:100329215 p38-beta; mitogen-activated protein kinase p38 b... 66.6 2e-11
ath:AT4G01370 ATMPK4; ATMPK4 (ARABIDOPSIS THALIANA MAP KINASE ... 66.2 2e-11
ath:AT5G19010 MPK16; MPK16; MAP kinase 65.9 3e-11
hsa:5594 MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2,... 65.9 3e-11
mmu:26413 Mapk1, 9030612K14Rik, AA407128, AU018647, C78273, ER... 65.9 3e-11
xla:494669 mapk11, p38-2, p38Beta, p38b, p38beta2, prkm11, sap... 65.5 4e-11
tgo:TGME49_033010 mitogen-activated protein kinase 2 (EC:2.7.1... 65.1 5e-11
cel:F42G8.3 pmk-2; P38 Map Kinase family member (pmk-2); K0444... 65.1 5e-11
ath:AT1G73670 ATMPK15; MAP kinase/ kinase 65.1 6e-11
dre:562552 fc89f08, wu:fc89f08; zgc:171775; K04441 p38 MAP kin... 64.7 6e-11
ath:AT2G46070 MPK12; MPK12 (MITOGEN-ACTIVATED PROTEIN KINASE 1... 63.9 1e-10
xla:379992 mapk14, MGC69036, csbp, mapk14a, mxi2, p38, p38-alp... 63.2 2e-10
> tgo:TGME49_007820 mitogen-activated protein kinase, putative
(EC:2.7.11.24); K08293 mitogen-activated protein kinase [EC:2.7.11.24]
Length=548
Score = 211 bits (536), Expect = 7e-55, Method: Compositional matrix adjust.
Identities = 97/128 (75%), Positives = 112/128 (87%), Gaps = 0/128 (0%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
ELILL ENYTEAIDVWSIGCIF+ELL+MI+ENV H DRGPLFPGSSCFPLSPDQK G+D
Sbjct 343 ELILLQENYTEAIDVWSIGCIFAELLNMIKENVAYHADRGPLFPGSSCFPLSPDQKAGND 402
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
+ TRGN+DQLNVIFN+LGTP+EE+IEALE +AK+YIR FP +E DLAERF ASS +
Sbjct 403 FKFHTRGNRDQLNVIFNILGTPSEEDIEALEKEDAKRYIRIFPKREGTDLAERFPASSAD 462
Query 121 ALNLLKRM 128
A++LLKRM
Sbjct 463 AIHLLKRM 470
> cpv:cgd2_4340 mitogen-activated protein kinase 2 ; K08293 mitogen-activated
protein kinase [EC:2.7.11.24]
Length=563
Score = 172 bits (436), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 79/129 (61%), Positives = 102/129 (79%), Gaps = 1/129 (0%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQK-NGD 59
ELILL ENYTEAID+WS+GCIF+ELL+M++ENV + DR PLFPGSSCFPLSPD K +
Sbjct 358 ELILLQENYTEAIDMWSVGCIFAELLNMLKENVAYYSDRSPLFPGSSCFPLSPDNKQQTE 417
Query 60 DSRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSP 119
D R + RGN+DQLN+IFNVLG+P +E+IE LE +AK+YIR F + DL RF +S
Sbjct 418 DYRFKIRGNRDQLNMIFNVLGSPLDEDIEVLEKEDAKRYIRIFSKRTGIDLQTRFPGASS 477
Query 120 EALNLLKRM 128
++++LLK+M
Sbjct 478 QSIDLLKKM 486
> bbo:BBOV_IV005520 23.m06268; mitogen-activated protein kinase;
K08293 mitogen-activated protein kinase [EC:2.7.11.24]
Length=584
Score = 152 bits (384), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 71/128 (55%), Positives = 93/128 (72%), Gaps = 4/128 (3%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
ELILL +NYT AIDVWS+GCIF+ELL+M++ N+ R PLFPG+SCFPLSPD KN D
Sbjct 385 ELILLQDNYTAAIDVWSVGCIFAELLNMVKGNMSDVSQRSPLFPGTSCFPLSPDNKNPSD 444
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
+ DQLN+IFNV+GTP EE+I A+ E ++YIR F P++ DL +RF + P
Sbjct 445 KAKE----HDQLNIIFNVIGTPCEEDIAAIIKPEVRRYIRMFHPRKGIDLYKRFKGTPPA 500
Query 121 ALNLLKRM 128
A+NLL++M
Sbjct 501 AVNLLQQM 508
> tpv:TP01_0359 mitogen-activated protein kinase 2; K08293 mitogen-activated
protein kinase [EC:2.7.11.24]
Length=642
Score = 152 bits (383), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 70/128 (54%), Positives = 96/128 (75%), Gaps = 3/128 (2%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
ELILL ENY+ A+D+WS+GCIF+ELL+M++ NV CDR PLFPGSSCFPLSPD KN +
Sbjct 442 ELILLQENYSFAVDIWSVGCIFAELLNMMKVNVSDPCDRSPLFPGSSCFPLSPDNKNANP 501
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
TR N DQLN+IFNVLGTP+EE+I ++ + ++Y++ F + DL +F +S E
Sbjct 502 R--TTREN-DQLNLIFNVLGTPSEEDINCIQKADVRRYVKFFAKRSFQDLRTKFKGASLE 558
Query 121 ALNLLKRM 128
+++LLK+M
Sbjct 559 SIDLLKKM 566
> pfa:PF11_0147 map-2; mitogen-activated protein kinase 2; K08293
mitogen-activated protein kinase [EC:2.7.11.24]
Length=508
Score = 143 bits (361), Expect = 1e-34, Method: Composition-based stats.
Identities = 63/128 (49%), Positives = 96/128 (75%), Gaps = 3/128 (2%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
ELILL ENYT +ID+WS GCIF+ELL+M++ ++ + +R PLFPGSSCFPLSPD +
Sbjct 302 ELILLQENYTNSIDIWSTGCIFAELLNMMKSHINNPTNRFPLFPGSSCFPLSPDH---NS 358
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
++ + N+DQLN+IFNV+GTP EE+++ + E +YI+ FP ++ DL++++S+ S E
Sbjct 359 KKVHEKSNRDQLNIIFNVIGTPPEEDLKCITKQEVIKYIKLFPTRDGIDLSKKYSSISKE 418
Query 121 ALNLLKRM 128
++LL+ M
Sbjct 419 GIDLLESM 426
> ath:AT2G18170 ATMPK7; ATMPK7 (ARABIDOPSIS THALIANA MAP KINASE
7); MAP kinase/ kinase; K08293 mitogen-activated protein kinase
[EC:2.7.11.24]
Length=368
Score = 78.6 bits (192), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 40/128 (31%), Positives = 69/128 (53%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
EL+L +NY +IDVWS+GCIF+E+L R P+FPG+ C
Sbjct 203 ELLLCCDNYGTSIDVWSVGCIFAEILG-----------RKPIFPGTECL----------- 240
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
+QL +I NV+G+ E +I +++ +A+++I++ P L+ + ++P
Sbjct 241 ---------NQLKLIINVVGSQQESDIRFIDNPKARRFIKSLPYSRGTHLSNLYPQANPL 291
Query 121 ALNLLKRM 128
A++LL+RM
Sbjct 292 AIDLLQRM 299
> ath:AT1G10210 ATMPK1; ATMPK1 (MITOGEN-ACTIVATED PROTEIN KINASE
1); MAP kinase/ kinase; K04371 extracellular signal-regulated
kinase 1/2 [EC:2.7.11.24]
Length=370
Score = 78.2 bits (191), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 42/128 (32%), Positives = 68/128 (53%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
EL+L +NY +IDVWS+GCIF+ELL R P+F G+ C
Sbjct 203 ELLLCCDNYGTSIDVWSVGCIFAELLG-----------RKPIFQGTECL----------- 240
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
+QL +I N+LG+ EE++E +++ +AK+YIR+ P L+ + +
Sbjct 241 ---------NQLKLIVNILGSQREEDLEFIDNPKAKRYIRSLPYSPGMSLSRLYPGAHVL 291
Query 121 ALNLLKRM 128
A++LL++M
Sbjct 292 AIDLLQKM 299
> ath:AT3G45640 ATMPK3; ATMPK3 (ARABIDOPSIS THALIANA MITOGEN-ACTIVATED
PROTEIN KINASE 3); MAP kinase/ kinase/ protein binding
/ protein kinase; K04371 extracellular signal-regulated
kinase 1/2 [EC:2.7.11.24]
Length=370
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 45/128 (35%), Positives = 67/128 (52%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
EL+L + +YT AIDVWS+GCIF EL+ +R PLFPG
Sbjct 208 ELLLNSSDYTAAIDVWSVGCIFMELM-----------NRKPLFPG--------------- 241
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
+ + Q+ ++ +LGTPTE ++ + +AK+YIR P LA+ FS +P
Sbjct 242 -----KDHVHQMRLLTELLGTPTESDLGFTHNEDAKRYIRQLPNFPRQPLAKLFSHVNPM 296
Query 121 ALNLLKRM 128
A++L+ RM
Sbjct 297 AIDLVDRM 304
> ath:AT1G59580 ATMPK2; ATMPK2 (ARABIDOPSIS THALIANA MITOGEN-ACTIVATED
PROTEIN KINASE HOMOLOG 2); MAP kinase/ kinase/ protein
kinase; K04371 extracellular signal-regulated kinase 1/2
[EC:2.7.11.24]
Length=376
Score = 76.3 bits (186), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 40/128 (31%), Positives = 68/128 (53%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
EL+L +NY +IDVWS+GCIF+ELL R P+FPG+ C
Sbjct 203 ELLLCCDNYGTSIDVWSVGCIFAELLG-----------RKPVFPGTECL----------- 240
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
+Q+ +I N+LG+ EE++E +++ +AK+YI + P + + ++
Sbjct 241 ---------NQIKLIINILGSQREEDLEFIDNPKAKRYIESLPYSPGISFSRLYPGANVL 291
Query 121 ALNLLKRM 128
A++LL++M
Sbjct 292 AIDLLQKM 299
> ath:AT4G36450 ATMPK14 (Mitogen-activated protein kinase 14);
MAP kinase/ kinase; K04371 extracellular signal-regulated kinase
1/2 [EC:2.7.11.24]
Length=361
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 37/128 (28%), Positives = 70/128 (54%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
EL+L +NY +IDVWS+GCIF+E+L R P+FPG+ C
Sbjct 200 ELLLCCDNYGTSIDVWSVGCIFAEILG-----------RKPIFPGTECL----------- 237
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
+QL +I NV+G+ + +++ +++ +A+++I++ P + + + ++P
Sbjct 238 ---------NQLKLIINVVGSQQDWDLQFIDNQKARRFIKSLPFSKGTHFSHIYPHANPL 288
Query 121 ALNLLKRM 128
A++LL+RM
Sbjct 289 AIDLLQRM 296
> ath:AT3G59790 ATMPK10; MAP kinase/ kinase; K04371 extracellular
signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=393
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 47/128 (36%), Positives = 68/128 (53%), Gaps = 32/128 (25%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
EL+L + +YT AIDVWS+GCIF E++ +R PLFPG DQ N
Sbjct 230 ELLLGSSDYTAAIDVWSVGCIFMEIM-----------NREPLFPGK-------DQVN--- 268
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
QL ++ ++GTP+EEE+ +L S AK+YIR P E+F P
Sbjct 269 ----------QLRLLLELIGTPSEEELGSL-SEYAKRYIRQLPTLPRQSFTEKFPNVPPL 317
Query 121 ALNLLKRM 128
A++L+++M
Sbjct 318 AIDLVEKM 325
> cel:B0218.3 pmk-1; P38 Map Kinase family member (pmk-1); K04441
p38 MAP kinase [EC:2.7.11.24]
Length=377
Score = 75.5 bits (184), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 42/128 (32%), Positives = 65/128 (50%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E++L +YT+ +DVWS+GCI +EL++ LFPGS
Sbjct 203 EIMLNWMHYTQTVDVWSVGCILAELIT-----------GKTLFPGSD------------- 238
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
+ DQL I +V GTP EE ++ + S EA+ YIR P D F+ ++P+
Sbjct 239 -------HIDQLTRIMSVTGTPDEEFLKKISSEEARNYIRNLPKMTRRDFKRLFAQATPQ 291
Query 121 ALNLLKRM 128
A++LL++M
Sbjct 292 AIDLLEKM 299
> xla:446599 mapk7, MGC81757, bmk1, erk4, prkm7; mitogen-activated
protein kinase 7; K04464 mitogen-activated protein kinase
7 [EC:2.7.11.24]
Length=925
Score = 74.3 bits (181), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 41/128 (32%), Positives = 65/128 (50%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
EL+L YT+AID+WS+GCIF+E+L R PLFPG++
Sbjct 226 ELMLSLHEYTQAIDMWSVGCIFAEMLG-----------RKPLFPGNNYL----------- 263
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
QL++I VLGTP+ + I A+ + + YI++ P ++ A + + +
Sbjct 264 ---------HQLHLIMTVLGTPSSQVIRAIGAERVRAYIQSLPSRQPVPWATLYPQAGKK 314
Query 121 ALNLLKRM 128
AL+LL +M
Sbjct 315 ALDLLSKM 322
> hsa:5595 MAPK3, ERK1, HS44KDAP, HUMKER1A, MGC20180, P44ERK1,
P44MAPK, PRKM3; mitogen-activated protein kinase 3 (EC:2.7.11.24);
K04371 extracellular signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=357
Score = 73.6 bits (179), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 40/128 (31%), Positives = 67/128 (52%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E++L ++ YT++ID+WS+GCI +E+LS P+FPG
Sbjct 214 EIMLNSKGYTKSIDIWSVGCILAEMLS-----------NRPIFPGKHYL----------- 251
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
DQLN I +LG+P++E++ + + +A+ Y+++ P K A+ F S +
Sbjct 252 ---------DQLNHILGILGSPSQEDLNCIINMKARNYLQSLPSKTKVAWAKLFPKSDSK 302
Query 121 ALNLLKRM 128
AL+LL RM
Sbjct 303 ALDLLDRM 310
> cel:F43C1.2 mpk-1; MAP Kinase family member (mpk-1); K04371
extracellular signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=444
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 39/128 (30%), Positives = 66/128 (51%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E++L ++ YT++IDVWS+GCI +E+LS PLFPG
Sbjct 268 EIMLNSKGYTKSIDVWSVGCILAEMLS-----------NRPLFPGKHYL----------- 305
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
DQLN+I V+G+P+ +++ + +++A+ Y+ + P K A + + P
Sbjct 306 ---------DQLNLILAVVGSPSNADLQCIINDKARSYLISLPHKPKQPWARLYPGADPR 356
Query 121 ALNLLKRM 128
AL+LL +M
Sbjct 357 ALDLLDKM 364
> mmu:26417 Mapk3, Erk-1, Erk1, Ert2, Esrk1, Mnk1, Mtap2k, Prkm3,
p44, p44erk1, p44mapk; mitogen-activated protein kinase
3 (EC:2.7.11.24); K04371 extracellular signal-regulated kinase
1/2 [EC:2.7.11.24]
Length=380
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 40/128 (31%), Positives = 67/128 (52%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E++L ++ YT++ID+WS+GCI +E+LS P+FPG
Sbjct 215 EIMLNSKGYTKSIDIWSVGCILAEMLS-----------NRPIFPGKHYL----------- 252
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
DQLN I +LG+P++E++ + + +A+ Y+++ P K A+ F S +
Sbjct 253 ---------DQLNHILGILGSPSQEDLNCIINMKARNYLQSLPSKTKVAWAKLFPKSDSK 303
Query 121 ALNLLKRM 128
AL+LL RM
Sbjct 304 ALDLLDRM 311
> dre:541323 mapk7, bmk1, erk5, wu:fb73b02, zgc:113111; mitogen-activated
protein kinase 7 (EC:2.7.1.-); K04464 mitogen-activated
protein kinase 7 [EC:2.7.11.24]
Length=862
Score = 72.8 bits (177), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 42/128 (32%), Positives = 61/128 (47%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
EL+L +Y+ AID+WS+GCIF E+L R +FPG +
Sbjct 257 ELMLSLHHYSLAIDLWSVGCIFGEMLG-----------RRQMFPGKNYV----------- 294
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
QL +I +VLGTP E + ++ S+ + Y+R+ P K LA + + P
Sbjct 295 ---------HQLQLILSVLGTPPESIVGSIGSDRVRSYVRSLPSKAPEPLAALYPQAEPS 345
Query 121 ALNLLKRM 128
ALNLL M
Sbjct 346 ALNLLAAM 353
> ath:AT2G43790 ATMPK6; ATMPK6 (ARABIDOPSIS THALIANA MAP KINASE
6); MAP kinase/ kinase; K14512 mitogen-activated protein kinase
6 [EC:2.7.11.24]
Length=395
Score = 72.4 bits (176), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 45/128 (35%), Positives = 66/128 (51%), Gaps = 32/128 (25%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
EL+L + +YT AIDVWS+GCIF EL+ DR PLFPG
Sbjct 233 ELLLNSSDYTAAIDVWSVGCIFMELM-----------DRKPLFPG--------------- 266
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
R + QL ++ ++GTP+EEE + AK+YIR PP + ++F P
Sbjct 267 -----RDHVHQLRLLMELIGTPSEEE-LEFLNENAKRYIRQLPPYPRQSITDKFPTVHPL 320
Query 121 ALNLLKRM 128
A++L+++M
Sbjct 321 AIDLIEKM 328
> hsa:5600 MAPK11, P38B, P38BETA2, PRKM11, SAPK2, SAPK2B, p38-2,
p38Beta; mitogen-activated protein kinase 11 (EC:2.7.11.24);
K04441 p38 MAP kinase [EC:2.7.11.24]
Length=364
Score = 70.9 bits (172), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 41/128 (32%), Positives = 61/128 (47%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E++L +Y + +D+WS+GCI +ELL LFPGS
Sbjct 192 EIMLNWMHYNQTVDIWSVGCIMAELLQ-----------GKALFPGSDYI----------- 229
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
DQL I V+GTP+ E + + S A+ YI++ PP DL+ F ++P
Sbjct 230 ---------DQLKRIMEVVGTPSPEVLAKISSEHARTYIQSLPPMPQKDLSSIFRGANPL 280
Query 121 ALNLLKRM 128
A++LL RM
Sbjct 281 AIDLLGRM 288
> hsa:100509694 mitogen-activated protein kinase 12-like
Length=357
Score = 70.9 bits (172), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 42/128 (32%), Positives = 64/128 (50%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E+IL YT+ +D+WS+GCI +E+++ G + F G D
Sbjct 185 EVILNWMRYTQTVDIWSVGCIMAEMIT-----------------GKTLF-------KGSD 220
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
+ DQL I V GTP E ++ L+S+EAK Y++ P E D A + +SP
Sbjct 221 -------HLDQLKEIMKVTGTPPAEFVQRLQSDEAKNYMKGLPELEKKDFASILTNASPL 273
Query 121 ALNLLKRM 128
A+NLL++M
Sbjct 274 AVNLLEKM 281
> hsa:6300 MAPK12, ERK3, ERK6, P38GAMMA, PRKM12, SAPK-3, SAPK3;
mitogen-activated protein kinase 12 (EC:2.7.11.24); K04441
p38 MAP kinase [EC:2.7.11.24]
Length=367
Score = 70.9 bits (172), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 42/128 (32%), Positives = 64/128 (50%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E+IL YT+ +D+WS+GCI +E+++ G + F G D
Sbjct 195 EVILNWMRYTQTVDIWSVGCIMAEMIT-----------------GKTLF-------KGSD 230
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
+ DQL I V GTP E ++ L+S+EAK Y++ P E D A + +SP
Sbjct 231 -------HLDQLKEIMKVTGTPPAEFVQRLQSDEAKNYMKGLPELEKKDFASILTNASPL 283
Query 121 ALNLLKRM 128
A+NLL++M
Sbjct 284 AVNLLEKM 291
> hsa:100509672 mitogen-activated protein kinase 12-like
Length=270
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 42/128 (32%), Positives = 62/128 (48%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E+IL YT+ +D+WS+GCI +E+++ LF GS
Sbjct 98 EVILNWMRYTQTVDIWSVGCIMAEMIT-----------GKTLFKGSDHL----------- 135
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
DQL I V GTP E ++ L+S+EAK Y++ P E D A + +SP
Sbjct 136 ---------DQLKEIMKVTGTPPAEFVQRLQSDEAKNYMKGLPELEKKDFASILTNASPL 186
Query 121 ALNLLKRM 128
A+NLL++M
Sbjct 187 AVNLLEKM 194
> mmu:19094 Mapk11, DKFZp586C1322, P38b, Prkm11, Sapk2, Sapk2b,
p38-2, p38beta, p38beta2; mitogen-activated protein kinase
11 (EC:2.7.11.24); K04441 p38 MAP kinase [EC:2.7.11.24]
Length=364
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 41/128 (32%), Positives = 63/128 (49%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E++L +Y + +D+WS+GCI +ELL G + FP G+D
Sbjct 192 EIMLNWMHYNQTVDIWSVGCIMAELLQ-----------------GKALFP-------GND 227
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
DQL I V+GTP+ E + + S A+ YI++ PP DL+ F ++P
Sbjct 228 Y-------IDQLKRIMEVVGTPSPEVLAKISSEHARTYIQSLPPMPQKDLSSVFHGANPL 280
Query 121 ALNLLKRM 128
A++LL RM
Sbjct 281 AIDLLGRM 288
> mmu:29857 Mapk12, AW123708, Erk6, P38gamma, Prkm12, Sapk3; mitogen-activated
protein kinase 12 (EC:2.7.11.24); K04441 p38
MAP kinase [EC:2.7.11.24]
Length=367
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 42/128 (32%), Positives = 63/128 (49%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E+IL YT+ +D+WS+GCI +E+++ G F G+D
Sbjct 195 EVILNWMRYTQTVDIWSVGCIMAEMIT-----------------GKILF-------KGND 230
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
+ DQL I + GTP E ++ L+S EAK Y+ P E D A + +SP+
Sbjct 231 -------HLDQLKEIMKITGTPPPEFVQKLQSAEAKNYMEGLPELEKKDFASVLTNASPQ 283
Query 121 ALNLLKRM 128
A+NLL+RM
Sbjct 284 AVNLLERM 291
> hsa:5598 MAPK7, BMK1, ERK4, ERK5, PRKM7; mitogen-activated protein
kinase 7 (EC:2.7.11.24); K04464 mitogen-activated protein
kinase 7 [EC:2.7.11.24]
Length=816
Score = 70.1 bits (170), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 41/128 (32%), Positives = 62/128 (48%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
EL+L YT+AID+WS+GCIF E+L+ R LFPG +
Sbjct 231 ELMLSLHEYTQAIDLWSVGCIFGEMLA-----------RRQLFPGKNYV----------- 268
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
QL +I VLGTP+ I+A+ + + YI++ PP++ + + +
Sbjct 269 ---------HQLQLIMMVLGTPSPAVIQAVGAERVRAYIQSLPPRQPVPWETVYPGADRQ 319
Query 121 ALNLLKRM 128
AL+LL RM
Sbjct 320 ALSLLGRM 327
> mmu:23939 Mapk7, BMK1, ERK5, Erk5-T, PRKM7; mitogen-activated
protein kinase 7 (EC:2.7.11.24); K04464 mitogen-activated
protein kinase 7 [EC:2.7.11.24]
Length=806
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 41/128 (32%), Positives = 62/128 (48%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
EL+L YT+AID+WS+GCIF E+L+ R LFPG +
Sbjct 231 ELMLSLHEYTQAIDLWSVGCIFGEMLA-----------RRQLFPGKNYV----------- 268
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
QL +I VLGTP+ I+A+ + + YI++ PP++ + + +
Sbjct 269 ---------HQLQLIMMVLGTPSPAVIQAVGAERVRAYIQSLPPRQPVPWETVYPGADRQ 319
Query 121 ALNLLKRM 128
AL+LL RM
Sbjct 320 ALSLLGRM 327
> sce:YBL016W FUS3, DAC2; Fus3p (EC:2.7.11.24); K04371 extracellular
signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=353
Score = 69.3 bits (168), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 40/129 (31%), Positives = 66/129 (51%), Gaps = 32/129 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E++L + Y+ A+DVWS GCI +EL R P+FPG
Sbjct 192 EVMLTSAKYSRAMDVWSCGCILAELFL-----------RRPIFPG--------------- 225
Query 61 SRLQTRGNKDQLNVIFNVLGTP-TEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSP 119
R + QL +IF ++GTP ++ ++ +ES A++YI++ P A L + F +P
Sbjct 226 -----RDYRHQLLLIFGIIGTPHSDNDLRCIESPRAREYIKSLPMYPAAPLEKMFPRVNP 280
Query 120 EALNLLKRM 128
+ ++LL+RM
Sbjct 281 KGIDLLQRM 289
> ath:AT1G01560 ATMPK11; MAP kinase/ kinase; K04371 extracellular
signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=369
Score = 68.9 bits (167), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 40/128 (31%), Positives = 61/128 (47%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
EL+L YT AID+WS+GCI E+++ R PLFPG
Sbjct 210 ELLLNCSEYTAAIDIWSVGCILGEIMT-----------REPLFPG--------------- 243
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
R QL +I ++G+P + + L S+ A++Y+R P + A RF S
Sbjct 244 -----RDYVQQLRLITELIGSPDDSSLGFLRSDNARRYVRQLPQYPRQNFAARFPNMSVN 298
Query 121 ALNLLKRM 128
A++LL++M
Sbjct 299 AVDLLQKM 306
> xla:398985 mapk1-a, MGC68934, erk, erk2, mapk, mapk1, mapk1a,
mpk1, xp42; mitogen-activated protein kinase 1 (EC:2.7.11.24);
K04371 extracellular signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=361
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 38/128 (29%), Positives = 65/128 (50%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E++L ++ YT++ID+WS+GCI +E+LS P+FPG
Sbjct 200 EIMLNSKGYTKSIDIWSVGCILAEMLS-----------NRPIFPGKHYL----------- 237
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
DQLN I +LG+P++E++ + + +A+ Y+ + P K F + P+
Sbjct 238 ---------DQLNHILGILGSPSQEDLNCIINLKARNYLLSLPHKNKVPWNRLFPNADPK 288
Query 121 ALNLLKRM 128
AL+LL +M
Sbjct 289 ALDLLDKM 296
> ath:AT1G07880 ATMPK13; MAP kinase/ kinase; K04371 extracellular
signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=363
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 40/128 (31%), Positives = 63/128 (49%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
EL+L + YT AID+WS+GCIF E+L R LFPG
Sbjct 203 ELLLNSSEYTGAIDIWSVGCIFMEILR-----------RETLFPG--------------- 236
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
+ QL +I +LG+P + +++ L S+ A++Y++ P + E+F SP
Sbjct 237 -----KDYVQQLKLITELLGSPDDSDLDFLRSDNARKYVKQLPHVQKQSFREKFPNISPM 291
Query 121 ALNLLKRM 128
AL+L ++M
Sbjct 292 ALDLAEKM 299
> xla:397785 mapk1-b, Xp42, erk, erk2, mapk, mapk1b, mpk1; mitogen-activated
protein kinase 1 (EC:2.7.11.24); K04371 extracellular
signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=361
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 38/128 (29%), Positives = 65/128 (50%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E++L ++ YT++ID+WS+GCI +E+LS P+FPG
Sbjct 200 EIMLNSKGYTKSIDIWSVGCILAEMLS-----------NRPIFPGKHYL----------- 237
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
DQLN I +LG+P++E++ + + +A+ Y+ + P K F + P+
Sbjct 238 ---------DQLNHILGILGSPSQEDLNCIINLKARNYLLSLPHKNKVPWNRLFPNADPK 288
Query 121 ALNLLKRM 128
AL+LL +M
Sbjct 289 ALDLLDKM 296
> ath:AT4G11330 ATMPK5; ATMPK5 (MAP KINASE 5); MAP kinase/ kinase;
K04371 extracellular signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=376
Score = 68.6 bits (166), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 39/128 (30%), Positives = 64/128 (50%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
EL+L + YT AIDVWS+GCIF+E+++ R PLFPG
Sbjct 213 ELLLNSSEYTSAIDVWSVGCIFAEIMT-----------REPLFPG--------------- 246
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
+ QL +I ++G+P +E L S A++Y++ P + + RF + +
Sbjct 247 -----KDYVHQLKLITELIGSPDGASLEFLRSANARKYVKELPKFPRQNFSARFPSMNST 301
Query 121 ALNLLKRM 128
A++LL++M
Sbjct 302 AIDLLEKM 309
> dre:399480 mapk3, ERK1, fi06b09, wu:fi06b09, zERK1; mitogen-activated
protein kinase 3 (EC:2.7.11.1); K04371 extracellular
signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=392
Score = 68.2 bits (165), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 38/128 (29%), Positives = 66/128 (51%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E++L ++ YT++ID+WS+GCI +E+LS P+FPG
Sbjct 228 EIMLNSKGYTKSIDIWSVGCILAEMLS-----------NRPIFPGKHYL----------- 265
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
DQLN I VLG+P+++++ + + +A+ Y+++ P K + F + +
Sbjct 266 ---------DQLNHILGVLGSPSQDDLNCIINMKARNYLQSLPQKPKIPWNKLFPKADNK 316
Query 121 ALNLLKRM 128
AL+LL RM
Sbjct 317 ALDLLDRM 324
> dre:100330818 mitogen-activated protein kinase 3-like; K04371
extracellular signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=392
Score = 68.2 bits (165), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 38/128 (29%), Positives = 66/128 (51%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E++L ++ YT++ID+WS+GCI +E+LS P+FPG
Sbjct 228 EIMLNSKGYTKSIDIWSVGCILAEMLS-----------NRPIFPGKHYL----------- 265
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
DQLN I VLG+P+++++ + + +A+ Y+++ P K + F + +
Sbjct 266 ---------DQLNHILGVLGSPSQDDLNCIINMKARNYLQSLPQKPKIPWNKLFPKADNK 316
Query 121 ALNLLKRM 128
AL+LL RM
Sbjct 317 ALDLLDRM 324
> dre:100002318 mapk13, im:7136778; mitogen-activated protein
kinase 13; K04441 p38 MAP kinase [EC:2.7.11.24]
Length=362
Score = 68.2 bits (165), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 43/128 (33%), Positives = 60/128 (46%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E+IL +YT+ +D+WS+GCI G +F G + F G D
Sbjct 189 EVILNWMHYTQTVDIWSVGCIM-----------------GEMFNGKTLF-------KGKD 224
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
DQL I V GTP E +E LES EAK Y+R+ P D + F +S +
Sbjct 225 Y-------MDQLTQIMKVAGTPGPEFVEKLESPEAKSYVRSLPHYPHRDFSTLFPRASKK 277
Query 121 ALNLLKRM 128
A+ LL++M
Sbjct 278 AVELLEKM 285
> dre:415185 zgc:86905; K04441 p38 MAP kinase [EC:2.7.11.24]
Length=361
Score = 68.2 bits (165), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 40/129 (31%), Positives = 64/129 (49%), Gaps = 33/129 (25%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGP-LFPGSSCFPLSPDQKNGD 59
E++L +Y + +D+WS+GCI ELL +G LFPG+
Sbjct 191 EIMLNWMHYNQTVDIWSVGCIMGELL------------KGKVLFPGNDYI---------- 228
Query 60 DSRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSP 119
DQL I V+GTPT + ++ + S A++YI++ P DL + F ++P
Sbjct 229 ----------DQLKRIMEVVGTPTPDVLKKISSEHAQKYIQSLPHMPQQDLGKIFRGANP 278
Query 120 EALNLLKRM 128
A++LLK+M
Sbjct 279 LAVDLLKKM 287
> xla:398592 mapk12, Xp38gamma, sapk3; mitogen-activated protein
kinase 12 (EC:2.7.11.24); K04441 p38 MAP kinase [EC:2.7.11.24]
Length=363
Score = 68.2 bits (165), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 36/128 (28%), Positives = 63/128 (49%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E+IL +YT+ +D+WS+GCI +E+ + PLF G+
Sbjct 195 EVILNWMHYTQTVDIWSVGCIMAEMYT-----------GRPLFKGNDHL----------- 232
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
+QL I + GTPT++ ++ L+S +AK YI++ P + D ++P
Sbjct 233 ---------NQLTEIMKITGTPTQDFVQKLQSTDAKNYIKSLPKVQKKDFGSLLRYANPL 283
Query 121 ALNLLKRM 128
A+N+L++M
Sbjct 284 AVNILEKM 291
> dre:360144 mapk1, zERK2; mitogen-activated protein kinase 1
(EC:2.7.11.24); K04371 extracellular signal-regulated kinase
1/2 [EC:2.7.11.24]
Length=369
Score = 67.8 bits (164), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 37/128 (28%), Positives = 65/128 (50%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E++L ++ YT++ID+WS+GCI +E+LS P+FPG
Sbjct 206 EIMLNSKGYTKSIDIWSVGCILAEMLS-----------NRPIFPGKHYL----------- 243
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
DQLN I +LG+P++E++ + + +A+ Y+ + P + F + P+
Sbjct 244 ---------DQLNHILGILGSPSQEDLNCIINIKARNYLLSLPLRSKVPWNRLFPNADPK 294
Query 121 ALNLLKRM 128
AL+LL +M
Sbjct 295 ALDLLDKM 302
> xla:100329215 p38-beta; mitogen-activated protein kinase p38
beta; K04441 p38 MAP kinase [EC:2.7.11.24]
Length=361
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 40/128 (31%), Positives = 62/128 (48%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E++L +Y + +D+WS+GCI +ELL G + FP G+D
Sbjct 191 EIMLNWMHYNQTVDIWSVGCIMAELLK-----------------GKALFP-------GND 226
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
DQL I V+GTP E + + S A++YI + P DL E F ++P
Sbjct 227 Y-------IDQLKRIMEVVGTPNSEFLMKISSEHARRYIESLPYMPQQDLKEVFHGANPL 279
Query 121 ALNLLKRM 128
A++LL++M
Sbjct 280 AIDLLEKM 287
> ath:AT4G01370 ATMPK4; ATMPK4 (ARABIDOPSIS THALIANA MAP KINASE
4); MAP kinase/ kinase; K04371 extracellular signal-regulated
kinase 1/2 [EC:2.7.11.24]
Length=376
Score = 66.2 bits (160), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 39/128 (30%), Positives = 60/128 (46%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
EL+L YT AID+WS+GCI E ++ R PLFPG
Sbjct 213 ELLLNCSEYTAAIDIWSVGCILGETMT-----------REPLFPG--------------- 246
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
+ QL +I ++G+P + + L S+ A++Y+R P + A RF S
Sbjct 247 -----KDYVHQLRLITELIGSPDDSSLGFLRSDNARRYVRQLPQYPRQNFAARFPNMSAG 301
Query 121 ALNLLKRM 128
A++LL++M
Sbjct 302 AVDLLEKM 309
> ath:AT5G19010 MPK16; MPK16; MAP kinase
Length=567
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 38/121 (31%), Positives = 61/121 (50%), Gaps = 31/121 (25%)
Query 8 NYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDDSRLQTRG 67
YT AID+WSIGCIF+ELL+ PLFPG +
Sbjct 207 KYTPAIDIWSIGCIFAELLTG-----------KPLFPGKNVV------------------ 237
Query 68 NKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPEALNLLKR 127
QL+++ ++LGTP+ E I + + +A++Y+ + K+ + +F + P AL LL++
Sbjct 238 --HQLDLMTDMLGTPSAEAIGRVRNEKARRYLSSMRKKKPIPFSHKFPHTDPLALRLLEK 295
Query 128 M 128
M
Sbjct 296 M 296
> hsa:5594 MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2,
p38, p40, p41, p41mapk; mitogen-activated protein kinase
1 (EC:2.7.11.24); K04371 extracellular signal-regulated kinase
1/2 [EC:2.7.11.24]
Length=360
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 37/128 (28%), Positives = 64/128 (50%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E++L ++ YT++ID+WS+GCI +E+LS P+FPG
Sbjct 197 EIMLNSKGYTKSIDIWSVGCILAEMLS-----------NRPIFPGKHYL----------- 234
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
DQLN I +LG+P++E++ + + +A+ Y+ + P K F + +
Sbjct 235 ---------DQLNHILGILGSPSQEDLNCIINLKARNYLLSLPHKNKVPWNRLFPNADSK 285
Query 121 ALNLLKRM 128
AL+LL +M
Sbjct 286 ALDLLDKM 293
> mmu:26413 Mapk1, 9030612K14Rik, AA407128, AU018647, C78273,
ERK, Erk2, MAPK2, PRKM2, Prkm1, p41mapk, p42mapk; mitogen-activated
protein kinase 1 (EC:2.7.11.24); K04371 extracellular
signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=358
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 37/128 (28%), Positives = 64/128 (50%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E++L ++ YT++ID+WS+GCI +E+LS P+FPG
Sbjct 195 EIMLNSKGYTKSIDIWSVGCILAEMLS-----------NRPIFPGKHYL----------- 232
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
DQLN I +LG+P++E++ + + +A+ Y+ + P K F + +
Sbjct 233 ---------DQLNHILGILGSPSQEDLNCIINLKARNYLLSLPHKNKVPWNRLFPNADSK 283
Query 121 ALNLLKRM 128
AL+LL +M
Sbjct 284 ALDLLDKM 291
> xla:494669 mapk11, p38-2, p38Beta, p38b, p38beta2, prkm11, sapk2,
sapk2b; mitogen-activated protein kinase 11; K04441 p38
MAP kinase [EC:2.7.11.24]
Length=361
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 40/128 (31%), Positives = 61/128 (47%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E++L +Y + +D+WS+GCI +ELL G + FP G+D
Sbjct 191 EIMLNWMHYNQTVDIWSVGCIMAELLK-----------------GKALFP-------GND 226
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
DQL I V GTP E + + S A++YI + P DL E F ++P
Sbjct 227 Y-------IDQLKRIMEVAGTPNSEFLMKISSEHARRYIESLPYMPHQDLKEVFRGANPL 279
Query 121 ALNLLKRM 128
A++LL++M
Sbjct 280 AIDLLEKM 287
> tgo:TGME49_033010 mitogen-activated protein kinase 2 (EC:2.7.11.24);
K08293 mitogen-activated protein kinase [EC:2.7.11.24]
Length=669
Score = 65.1 bits (157), Expect = 5e-11, Method: Composition-based stats.
Identities = 38/128 (29%), Positives = 65/128 (50%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E++L + +YT+ +D+WS+GCI ELLS P+FPG+S
Sbjct 190 EILLGSTSYTKGVDMWSLGCILGELLS-----------GRPIFPGTSTM----------- 227
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
+QL I + G P+ E+++A++S A + + P + + + F +SPE
Sbjct 228 ---------NQLERIMTLTGRPSPEDVDAVKSPFAATMMESLPLGKVKNFKDAFPNASPE 278
Query 121 ALNLLKRM 128
AL+LLK++
Sbjct 279 ALDLLKQL 286
> cel:F42G8.3 pmk-2; P38 Map Kinase family member (pmk-2); K04441
p38 MAP kinase [EC:2.7.11.24]
Length=419
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 42/121 (34%), Positives = 58/121 (47%), Gaps = 31/121 (25%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E++L +YT+ +DVWS+GCI +EL+S PLFPG DD
Sbjct 234 EIMLNWMHYTQTVDVWSVGCILAELVS-----------GRPLFPG-------------DD 269
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
+ DQL I +V+GTP EE ++S EA+ YI+ P D F +SP
Sbjct 270 -------HIDQLTKIMSVVGTPKEEFWSKIQSEEARNYIKNRSPIIRQDFVTLFPMASPY 322
Query 121 A 121
A
Sbjct 323 A 323
> ath:AT1G73670 ATMPK15; MAP kinase/ kinase
Length=576
Score = 65.1 bits (157), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 36/120 (30%), Positives = 59/120 (49%), Gaps = 31/120 (25%)
Query 9 YTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDDSRLQTRGN 68
YT AID+WS+GCIF+E+L PLFPG +
Sbjct 273 YTPAIDIWSVGCIFAEMLLG-----------KPLFPGKNVV------------------- 302
Query 69 KDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPEALNLLKRM 128
QL+++ + LGTP E I + +++A++Y+ K+ +++F + P AL LL+R+
Sbjct 303 -HQLDIMTDFLGTPPPEAISKIRNDKARRYLGNMRKKQPVPFSKKFPKADPSALRLLERL 361
> dre:562552 fc89f08, wu:fc89f08; zgc:171775; K04441 p38 MAP kinase
[EC:2.7.11.24]
Length=359
Score = 64.7 bits (156), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 40/128 (31%), Positives = 59/128 (46%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E+I +YT+ +DVW+ GCI +E MI V LFPGS
Sbjct 193 EVIFNWMHYTQTVDVWTAGCILAE---MITGEV--------LFPGSDSI----------- 230
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
DQL I N+ GTP + ++S +A+ Y+R+ P ++ E FS P
Sbjct 231 ---------DQLKKILNLTGTPNSTLVLKMQSKDAQSYVRSLPVQKKKAFKEVFSGMDPN 281
Query 121 ALNLLKRM 128
A++LL+ M
Sbjct 282 AIDLLEGM 289
> ath:AT2G46070 MPK12; MPK12 (MITOGEN-ACTIVATED PROTEIN KINASE
12); MAP kinase/ kinase; K04371 extracellular signal-regulated
kinase 1/2 [EC:2.7.11.24]
Length=372
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 38/128 (29%), Positives = 57/128 (44%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
EL+L YT AID+WS+GCI E+++ PLFPG
Sbjct 211 ELLLNCSEYTAAIDIWSVGCILGEIMTG-----------QPLFPG--------------- 244
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
+ QL +I ++G+P + L S+ A++Y+R P A RF
Sbjct 245 -----KDYVHQLRLITELVGSPDNSSLGFLRSDNARRYVRQLPRYPKQQFAARFPKMPTT 299
Query 121 ALNLLKRM 128
A++LL+RM
Sbjct 300 AIDLLERM 307
> xla:379992 mapk14, MGC69036, csbp, mapk14a, mxi2, p38, p38-alpha,
sapk2, sapk2a; mitogen-activated protein kinase 14 (EC:2.7.11.24);
K04441 p38 MAP kinase [EC:2.7.11.24]
Length=361
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 36/128 (28%), Positives = 65/128 (50%), Gaps = 31/128 (24%)
Query 1 ELILLAENYTEAIDVWSIGCIFSELLSMIEENVESHCDRGPLFPGSSCFPLSPDQKNGDD 60
E++L +Y + +D+WS+GCI +ELL+ G + FP G D
Sbjct 193 EIMLNWMHYNQTVDIWSVGCIMAELLT-----------------GRTLFP-------GTD 228
Query 61 SRLQTRGNKDQLNVIFNVLGTPTEEEIEALESNEAKQYIRTFPPKEAHDLAERFSASSPE 120
+ DQL +I ++GTP E ++ + S A+ YI++ P + + F ++P+
Sbjct 229 -------HIDQLKLILRLVGTPEPELLQKISSEAARNYIQSLPYMPKMNFEDVFLGANPQ 281
Query 121 ALNLLKRM 128
A++LL++M
Sbjct 282 AVDLLEKM 289
Lambda K H
0.314 0.133 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2049573556
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40