bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2542_orf1
Length=135
Score E
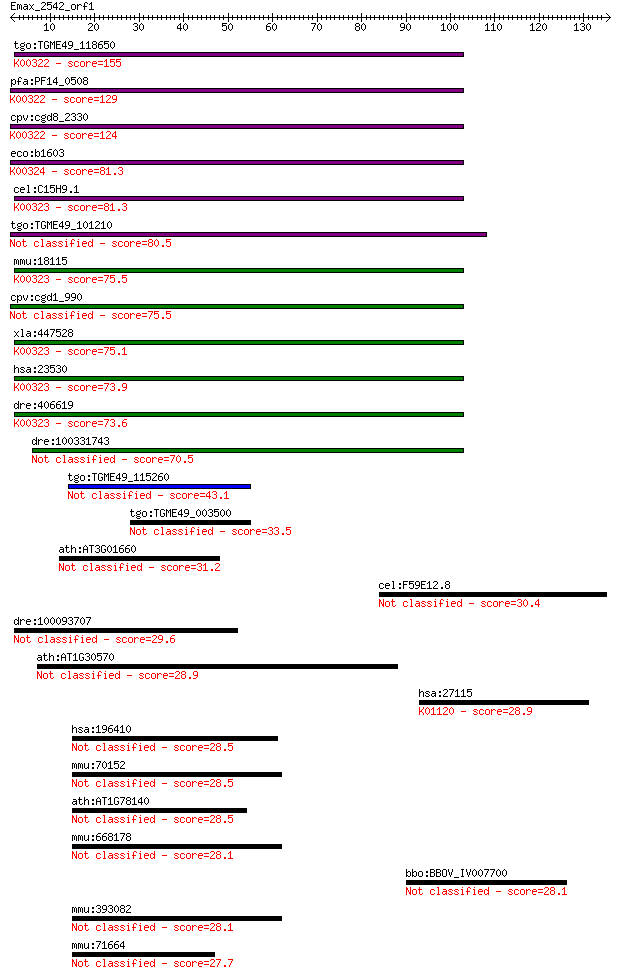
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00... 155 2e-38
pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative (... 129 2e-30
cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alp... 124 1e-28
eco:b1603 pntA, ECK1598, JW1595; pyridine nucleotide transhydr... 81.3 7e-16
cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase fa... 81.3 7e-16
tgo:TGME49_101210 NAD(P) transhydrogenase, alpha subunit, puta... 80.5 1e-15
mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide... 75.5 4e-14
cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alph... 75.5 5e-14
xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydroge... 75.1 5e-14
hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide t... 73.9 1e-13
dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamid... 73.6 2e-13
dre:100331743 Nicotinamide Nucleotide Transhydrogenase family ... 70.5 1e-12
tgo:TGME49_115260 alanine dehydrogenase, putative (EC:1.4.1.1) 43.1 2e-04
tgo:TGME49_003500 alanine dehydrogenase, putative (EC:1.4.1.1) 33.5 0.20
ath:AT3G01660 methyltransferase 31.2 1.0
cel:F59E12.8 hypothetical protein 30.4 1.7
dre:100093707 wu:fj43a03; zgc:165628 29.6 3.0
ath:AT1G30570 protein kinase family protein 28.9 4.6
hsa:27115 PDE7B, MGC88256, bA472E5.1; phosphodiesterase 7B (EC... 28.9 5.0
hsa:196410 METTL7B, MGC17301; methyltransferase like 7B 28.5 5.6
mmu:70152 Mettl7a1, 2210414H16Rik, 3300001H21Rik, Aam-B, Mettl... 28.5 6.1
ath:AT1G78140 methyltransferase-related 28.5 6.7
mmu:668178 Gm10035, Ubie3; predicted gene 10035 28.1
bbo:BBOV_IV007700 23.m06563; hypothetical protein 28.1 8.4
mmu:393082 Mettl7a2, Gm921, UbiE2, Ubie; methyltransferase lik... 28.1 8.4
mmu:71664 Mettl7b, 0610006F02Rik, AI266817; methyltransferase ... 27.7 9.5
> tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00322
NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1013
Score = 155 bits (393), Expect = 2e-38, Method: Composition-based stats.
Identities = 67/101 (66%), Positives = 85/101 (84%), Gaps = 0/101 (0%)
Query 2 AQRELIANTIKHCDVVICTAAIHGKPSPKLISRDMLRTMKPGSVIVDLATEFGDVRAGWG 61
AQ+++++ + + DV+ICTAAIHGKPSPKLIS DML TM+PGSV++DLATEFGD R+ WG
Sbjct 734 AQKQMLSRVVPNVDVIICTAAIHGKPSPKLISNDMLATMRPGSVVIDLATEFGDRRSNWG 793
Query 62 GNVEVAPKDDQIVVDGITVIGRRRIETRMPVQASELFSMNM 102
GNVE +P D + + G+T+IGR RIET+MP QASELFSMNM
Sbjct 794 GNVEGSPTDGETQIHGVTIIGRSRIETQMPTQASELFSMNM 834
> pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative
(EC:1.6.1.2); K00322 NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1176
Score = 129 bits (325), Expect = 2e-30, Method: Composition-based stats.
Identities = 54/102 (52%), Positives = 78/102 (76%), Gaps = 0/102 (0%)
Query 1 RAQRELIANTIKHCDVVICTAAIHGKPSPKLISRDMLRTMKPGSVIVDLATEFGDVRAGW 60
+ Q L IK CD++IC+A+I GK SPKL++ +M++ MKPGSV VDL+TEFGD + W
Sbjct 880 KVQSNLFKKIIKKCDILICSASIPGKTSPKLVTTEMIKLMKPGSVAVDLSTEFGDKKNNW 939
Query 61 GGNVEVAPKDDQIVVDGITVIGRRRIETRMPVQASELFSMNM 102
GGN+E + ++ I+++G+ V+GR +IE MP+QAS+LFSMNM
Sbjct 940 GGNIECSQSNENILINGVHVLGRDKIERNMPMQASDLFSMNM 981
> cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, possible signal peptide
plus 12 transmembrane regions ; K00322 NAD(P) transhydrogenase
[EC:1.6.1.1]
Length=1143
Score = 124 bits (310), Expect = 1e-28, Method: Composition-based stats.
Identities = 59/105 (56%), Positives = 77/105 (73%), Gaps = 4/105 (3%)
Query 1 RAQRELIANTIKHCDVVICTAAIHGKPSPKLISRDMLRTMKPGSVIVDLATEFGDVRAGW 60
+AQ +L + I+ CDVVI TA I GKPSPK+I+R+M+ +MKPGSVIVD+A E D +GW
Sbjct 823 KAQSKLYSKMIRSCDVVITTALIPGKPSPKIITREMVNSMKPGSVIVDMAAEMADTASGW 882
Query 61 GGNVEVAPKDDQIVVD---GITVIGRRRIETRMPVQASELFSMNM 102
GGN E+ K DQI +D G+T+IG + + MP QASELFSMN+
Sbjct 883 GGNCEIT-KKDQIYLDEKSGVTIIGLTNLPSTMPSQASELFSMNV 926
> eco:b1603 pntA, ECK1598, JW1595; pyridine nucleotide transhydrogenase,
alpha subunit (EC:1.6.1.2); K00324 NAD(P) transhydrogenase
subunit alpha [EC:1.6.1.2]
Length=510
Score = 81.3 bits (199), Expect = 7e-16, Method: Composition-based stats.
Identities = 43/103 (41%), Positives = 63/103 (61%), Gaps = 8/103 (7%)
Query 1 RAQRELIANTIKHCDVVICTAAIHGKPSPKLISRDMLRTMKPGSVIVDLATEFGDVRAGW 60
+A+ EL A K D+++ TA I GKP+PKLI+R+M+ +MK GSVIVDLA +
Sbjct 236 KAEMELFAAQAKEVDIIVTTALIPGKPAPKLITREMVDSMKAGSVIVDLAAQN------- 288
Query 61 GGNVE-VAPKDDQIVVDGITVIGRRRIETRMPVQASELFSMNM 102
GGN E P + +G+ VIG + R+P Q+S+L+ N+
Sbjct 289 GGNCEYTVPGEIFTTENGVKVIGYTDLPGRLPTQSSQLYGTNL 331
> cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase
family member (nnt-1); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1041
Score = 81.3 bits (199), Expect = 7e-16, Method: Composition-based stats.
Identities = 42/101 (41%), Positives = 63/101 (62%), Gaps = 7/101 (6%)
Query 2 AQRELIANTIKHCDVVICTAAIHGKPSPKLISRDMLRTMKPGSVIVDLATEFGDVRAGWG 61
A+ +L A+ K D++I TA I GK +P LI+ +M+++MKPGSV+VDLA E G
Sbjct 265 AEMKLFADQCKDVDIIITTALIPGKKAPILITEEMIKSMKPGSVVVDLAAES-------G 317
Query 62 GNVEVAPKDDQIVVDGITVIGRRRIETRMPVQASELFSMNM 102
GN+ + V G+T IG + +R+P Q+SEL+S N+
Sbjct 318 GNIATTRPGEVYVKHGVTHIGFTDLPSRLPTQSSELYSNNI 358
> tgo:TGME49_101210 NAD(P) transhydrogenase, alpha subunit, putative
(EC:1.6.1.2)
Length=1165
Score = 80.5 bits (197), Expect = 1e-15, Method: Composition-based stats.
Identities = 45/108 (41%), Positives = 67/108 (62%), Gaps = 8/108 (7%)
Query 1 RAQRELIANTIKHCDVVICTAAIHGKPSPKLISRDMLRTMKPGSVIVDLATEFGDVRAGW 60
R + EL A + D++I TA++ G+P+PKLI ++ + TMK GSVIVDLA A
Sbjct 837 RKEMELFAEQCREIDILITTASLPGRPAPKLIKKEHVDTMKAGSVIVDLA-------APT 889
Query 61 GGNVEVAPKDDQIVVDG-ITVIGRRRIETRMPVQASELFSMNMWRDFE 107
GGNVEV + + +G +TV+G + +RM QASE+F+ N++ E
Sbjct 890 GGNVEVTRPGETYLYNGKVTVVGWTDLPSRMAPQASEMFARNIFNLLE 937
> mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 75.5 bits (184), Expect = 4e-14, Method: Composition-based stats.
Identities = 43/101 (42%), Positives = 58/101 (57%), Gaps = 7/101 (6%)
Query 2 AQRELIANTIKHCDVVICTAAIHGKPSPKLISRDMLRTMKPGSVIVDLATEFGDVRAGWG 61
A+ +L A K D++I TA I GK +P L S++M+ +MK GSV+VDLA E G
Sbjct 299 AEMKLFAQQCKEVDILISTALIPGKKAPVLFSKEMIESMKEGSVVVDLAAEA-------G 351
Query 62 GNVEVAPKDDQIVVDGITVIGRRRIETRMPVQASELFSMNM 102
GN E + V GIT IG + +RM QAS L+S N+
Sbjct 352 GNFETTKPGELYVHKGITHIGYTDLPSRMATQASTLYSNNI 392
> cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, 12 transmembrane domain
(EC:1.6.1.2)
Length=1147
Score = 75.5 bits (184), Expect = 5e-14, Method: Composition-based stats.
Identities = 44/104 (42%), Positives = 61/104 (58%), Gaps = 9/104 (8%)
Query 1 RAQRELIANTIKHCDVVICTAAIHGKPSPKLISRDMLRTMKPGSVIVDLATEFGDVRAGW 60
+ QR+LI + D+VI +A G+ P LI+++ +R MK GSVIVDL +EF
Sbjct 828 KKQRDLIEKYLCISDIVITSACKPGEECPILITKEAVRKMKSGSVIVDLCSEF------- 880
Query 61 GGNVEVAPKDDQI--VVDGITVIGRRRIETRMPVQASELFSMNM 102
GGN E+ KD V G T+IG+ MP+Q+SELFS N+
Sbjct 881 GGNCELTQKDRTFSDVQSGTTIIGKCNYVFSMPLQSSELFSGNL 924
> xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydrogenase
(EC:1.6.1.2); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1086
Score = 75.1 bits (183), Expect = 5e-14, Method: Composition-based stats.
Identities = 42/101 (41%), Positives = 57/101 (56%), Gaps = 7/101 (6%)
Query 2 AQRELIANTIKHCDVVICTAAIHGKPSPKLISRDMLRTMKPGSVIVDLATEFGDVRAGWG 61
A+ +L A + D+++ TA I GK +P L +DM+ MK GSV+VDLA E G
Sbjct 299 AEMKLFAKQCQDVDIIVTTALIPGKTAPILFRKDMIELMKEGSVVVDLAAEA-------G 351
Query 62 GNVEVAPKDDQIVVDGITVIGRRRIETRMPVQASELFSMNM 102
GN+E D V G+T IG I +RM QAS L+S N+
Sbjct 352 GNIETTKPGDIYVHKGVTHIGYTDIPSRMASQASTLYSNNI 392
> hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide
transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 73.9 bits (180), Expect = 1e-13, Method: Composition-based stats.
Identities = 41/101 (40%), Positives = 58/101 (57%), Gaps = 7/101 (6%)
Query 2 AQRELIANTIKHCDVVICTAAIHGKPSPKLISRDMLRTMKPGSVIVDLATEFGDVRAGWG 61
A+ +L A K D++I TA I GK +P L +++M+ +MK GSV+VDLA E G
Sbjct 299 AEMKLFAQQCKEVDILISTALIPGKKAPVLFNKEMIESMKEGSVVVDLAAEA-------G 351
Query 62 GNVEVAPKDDQIVVDGITVIGRRRIETRMPVQASELFSMNM 102
GN E + + GIT IG + +RM QAS L+S N+
Sbjct 352 GNFETTKPGELYIHKGITHIGYTDLPSRMATQASTLYSNNI 392
> dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P)
transhydrogenase [EC:1.6.1.2]
Length=1079
Score = 73.6 bits (179), Expect = 2e-13, Method: Composition-based stats.
Identities = 40/101 (39%), Positives = 59/101 (58%), Gaps = 7/101 (6%)
Query 2 AQRELIANTIKHCDVVICTAAIHGKPSPKLISRDMLRTMKPGSVIVDLATEFGDVRAGWG 61
A+ +L A D++I TA I G+ +P LI+++M+ TMK GSV+VDLA E G
Sbjct 295 AEMKLFAKQCLDVDIIITTALIPGRKAPVLITKEMVETMKDGSVVVDLAAEA-------G 347
Query 62 GNVEVAPKDDQIVVDGITVIGRRRIETRMPVQASELFSMNM 102
GN+E + V G+ +G I +R+P QAS L+S N+
Sbjct 348 GNIETTVPGELSVHKGVIHVGYTDIPSRLPTQASTLYSNNI 388
> dre:100331743 Nicotinamide Nucleotide Transhydrogenase family
member (nnt-1)-like
Length=518
Score = 70.5 bits (171), Expect = 1e-12, Method: Composition-based stats.
Identities = 39/97 (40%), Positives = 55/97 (56%), Gaps = 7/97 (7%)
Query 6 LIANTIKHCDVVICTAAIHGKPSPKLISRDMLRTMKPGSVIVDLATEFGDVRAGWGGNVE 65
L A + D++I TA GK +P LI R+M+ +M+ GSV+VDLA E GGN+E
Sbjct 3 LFAKQCRDVDIIISTALNPGKRAPVLIKREMVESMRDGSVVVDLAAEA-------GGNIE 55
Query 66 VAPKDDQIVVDGITVIGRRRIETRMPVQASELFSMNM 102
+ V G+T IG + +RM QAS L+S N+
Sbjct 56 TTKPGELYVHQGVTHIGYTDLPSRMATQASSLYSNNI 92
> tgo:TGME49_115260 alanine dehydrogenase, putative (EC:1.4.1.1)
Length=390
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 17/41 (41%), Positives = 30/41 (73%), Gaps = 0/41 (0%)
Query 14 CDVVICTAAIHGKPSPKLISRDMLRTMKPGSVIVDLATEFG 54
CD+++ + G+ +PK++ R+ L+ MKPGSVIVD++ + G
Sbjct 231 CDLLVGAVLLPGRKAPKIVRREHLKRMKPGSVIVDVSIDHG 271
> tgo:TGME49_003500 alanine dehydrogenase, putative (EC:1.4.1.1)
Length=462
Score = 33.5 bits (75), Expect = 0.20, Method: Composition-based stats.
Identities = 14/27 (51%), Positives = 19/27 (70%), Gaps = 0/27 (0%)
Query 28 SPKLISRDMLRTMKPGSVIVDLATEFG 54
+P L+ ++ + MKPGSVIVDLA G
Sbjct 294 APMLVKKEHVAMMKPGSVIVDLAVSRG 320
> ath:AT3G01660 methyltransferase
Length=273
Score = 31.2 bits (69), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 10/36 (27%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 12 KHCDVVICTAAIHGKPSPKLISRDMLRTMKPGSVIV 47
K+ D V+C+ + P+ + ++ R +KPG V++
Sbjct 153 KYFDAVLCSVGVQYLQQPEKVFAEVYRVLKPGGVLI 188
> cel:F59E12.8 hypothetical protein
Length=412
Score = 30.4 bits (67), Expect = 1.7, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 26/53 (49%), Gaps = 4/53 (7%)
Query 84 RRIETRMPVQASELFSMNMW--RDFEICLFQHAFSPRSCLPVLCRLFHTLSKR 134
+RI ++ + ELF ++W RDF +C A+ R P L L + KR
Sbjct 243 QRISSKFSIWTEELFHSDIWLIRDFPVC--PAAYIVRPTFPYLDELNEMIIKR 293
> dre:100093707 wu:fj43a03; zgc:165628
Length=208
Score = 29.6 bits (65), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 24/50 (48%), Gaps = 1/50 (2%)
Query 2 AQRELIANTIKHCDVVICTAAIHGKPSPKLISRDMLRTMKPGSVIVDLAT 51
+QRE+I +H D + A +H P P L+ M + PG DL T
Sbjct 13 SQREIITPEAEHVDAGLAVADLHSHPHP-LLDPSMPGPLPPGLGDHDLLT 61
> ath:AT1G30570 protein kinase family protein
Length=849
Score = 28.9 bits (63), Expect = 4.6, Method: Composition-based stats.
Identities = 19/81 (23%), Positives = 39/81 (48%), Gaps = 4/81 (4%)
Query 7 IANTIKHCDVVICTAAIHGKPSPKLISRDMLRTMKPGSVIVDLATEFGDVRAGWGGNVEV 66
IA I H ++++ + H + L+ +L T PG +++ E G G+ +E+
Sbjct 141 IAGEIAHKNLILESTG-HNATASSLVKEFLLPT-GPGKLVLSFIPEKGSF--GFVNAIEI 196
Query 67 APKDDQIVVDGITVIGRRRIE 87
DD++ + +T +G +E
Sbjct 197 VSVDDKLFKESVTKVGGSEVE 217
> hsa:27115 PDE7B, MGC88256, bA472E5.1; phosphodiesterase 7B (EC:3.1.4.17);
K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=450
Score = 28.9 bits (63), Expect = 5.0, Method: Composition-based stats.
Identities = 16/39 (41%), Positives = 23/39 (58%), Gaps = 2/39 (5%)
Query 93 QASELFS-MNMWRDFEICLFQHAFSPRSCLPVLCRLFHT 130
QA + S + MW DF+I LF + S + +LC LF+T
Sbjct 104 QARHMLSKVGMW-DFDIFLFDRLTNGNSLVTLLCHLFNT 141
> hsa:196410 METTL7B, MGC17301; methyltransferase like 7B
Length=244
Score = 28.5 bits (62), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 14/49 (28%), Positives = 26/49 (53%), Gaps = 3/49 (6%)
Query 15 DVVICTAAIHGKPSPKLISRDMLRTMKPGSVIV---DLATEFGDVRAGW 60
DVV+CT + SP+ + +++ R ++PG V+ +A +G W
Sbjct 139 DVVVCTLVLCSVQSPRKVLQEVRRVLRPGGVLFFWEHVAEPYGSWAFMW 187
> mmu:70152 Mettl7a1, 2210414H16Rik, 3300001H21Rik, Aam-B, Mettl7a,
UbiE1; methyltransferase like 7A1
Length=244
Score = 28.5 bits (62), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 22/47 (46%), Gaps = 1/47 (2%)
Query 15 DVVICTAAIHGKPSPKLISRDMLRTMKPGSVIVDLATEFGDVRAGWG 61
DVV+CT + + + I R++ R +KPG D R+ W
Sbjct 139 DVVVCTLVLCSVKNQEKILREVCRVLKPGGAFY-FMEHVADERSTWN 184
> ath:AT1G78140 methyltransferase-related
Length=355
Score = 28.5 bits (62), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 21/39 (53%), Gaps = 2/39 (5%)
Query 15 DVVICTAAIHGKPSPKLISRDMLRTMKPGSVIVDLATEF 53
D V AA+H PSP ++ R ++PG V V AT F
Sbjct 256 DAVHAGAALHCWPSPSSAVAEISRVLRPGGVFV--ATTF 292
> mmu:668178 Gm10035, Ubie3; predicted gene 10035
Length=244
Score = 28.1 bits (61), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 22/47 (46%), Gaps = 1/47 (2%)
Query 15 DVVICTAAIHGKPSPKLISRDMLRTMKPGSVIVDLATEFGDVRAGWG 61
DVV+CT + + + I R++ R +KPG D R+ W
Sbjct 139 DVVVCTLVLCSVKNQEKILREVCRVLKPGGAFY-FIDHVADERSTWN 184
> bbo:BBOV_IV007700 23.m06563; hypothetical protein
Length=202
Score = 28.1 bits (61), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 10/36 (27%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 90 MPVQASELFSMNMWRDFEICLFQHAFSPRSCLPVLC 125
+P+ +++FS+N D ++C+ + F+ +P LC
Sbjct 51 VPLTMAKIFSLNDIYDAQVCVLNNIFAENKDIPKLC 86
> mmu:393082 Mettl7a2, Gm921, UbiE2, Ubie; methyltransferase like
7A2
Length=244
Score = 28.1 bits (61), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 22/47 (46%), Gaps = 1/47 (2%)
Query 15 DVVICTAAIHGKPSPKLISRDMLRTMKPGSVIVDLATEFGDVRAGWG 61
DVV+CT + + + I R++ R +KPG D R+ W
Sbjct 139 DVVVCTLVLCSVKNQEKILREVCRVLKPGGAFY-FIDHVADERSTWN 184
> mmu:71664 Mettl7b, 0610006F02Rik, AI266817; methyltransferase
like 7B
Length=244
Score = 27.7 bits (60), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 10/32 (31%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 15 DVVICTAAIHGKPSPKLISRDMLRTMKPGSVI 46
DVV+CT + SP+ + +++ R ++PG ++
Sbjct 139 DVVVCTLVLCSVQSPRKVLQEVQRVLRPGGLL 170
Lambda K H
0.327 0.139 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2296762580
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40