bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2500_orf1
Length=86
Score E
Sequences producing significant alignments: (Bits) Value
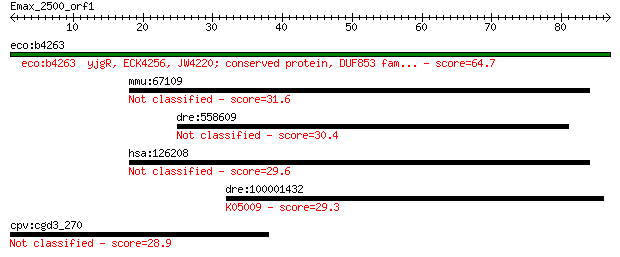
eco:b4263 yjgR, ECK4256, JW4220; conserved protein, DUF853 fam... 64.7 7e-11
mmu:67109 Zfp787, 2210018M03Rik, Znf787; zinc finger protein 787 31.6
dre:558609 fj65a11, wu:fj65a11; si:dkey-73n10.1 30.4 1.6
hsa:126208 ZNF787, TIP20; zinc finger protein 787 29.6
dre:100001432 eKir-like; K05009 potassium inwardly-rectifying ... 29.3 3.4
cpv:cgd3_270 hypothetical protein 28.9 4.6
> eco:b4263 yjgR, ECK4256, JW4220; conserved protein, DUF853 family
with NTPase fold; K06915
Length=500
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 33/86 (38%), Positives = 54/86 (62%), Gaps = 0/86 (0%)
Query 1 FKVADREGLLLLDLKDLKALLNHLRYHPELLGEDAALMTTGSSLALLRRLAVLEQQGAEA 60
F++AD +GLLLLD KDL+A+ ++ + + +++ S A+ R L LEQQGA
Sbjct 138 FRIADDQGLLLLDFKDLRAITQYIGDNAKSFQNQYGNISSASVGAIQRGLLSLEQQGAAH 197
Query 61 LFGEPALQLEDILQPANDGRGRIHLL 86
FGEP L ++D ++ +G+G I++L
Sbjct 198 FFGEPMLDIKDWMRTDANGKGVINIL 223
> mmu:67109 Zfp787, 2210018M03Rik, Znf787; zinc finger protein
787
Length=381
Score = 31.6 bits (70), Expect = 0.64, Method: Composition-based stats.
Identities = 25/66 (37%), Positives = 30/66 (45%), Gaps = 12/66 (18%)
Query 18 KALLNHLRYHPELLGEDAALMTTGSSLALLRRLAVLEQQGAEALFGEPALQLEDILQPAN 77
K+L HLR HPEL G A +S +RR E+ A A GE A+ P
Sbjct 191 KSLARHLRLHPELSGPGVAAKVLAAS---VRRAKAPEE--ATAADGEIAI-------PVG 238
Query 78 DGRGRI 83
DG G I
Sbjct 239 DGEGII 244
> dre:558609 fj65a11, wu:fj65a11; si:dkey-73n10.1
Length=1277
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 20/56 (35%), Positives = 27/56 (48%), Gaps = 8/56 (14%)
Query 25 RYHPELLGEDAALMTTGSSLALLRRLAVLEQQGAEALFGEPALQLEDILQPANDGR 80
R P + ED A A RLA L ++G EAL + LQLE ++ DG+
Sbjct 212 RMSPHIYDEDWA--------AHYSRLAYLHKKGQEALAQQRGLQLEHGVEGIKDGK 259
> hsa:126208 ZNF787, TIP20; zinc finger protein 787
Length=382
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 25/66 (37%), Positives = 30/66 (45%), Gaps = 12/66 (18%)
Query 18 KALLNHLRYHPELLGEDAALMTTGSSLALLRRLAVLEQQGAEALFGEPALQLEDILQPAN 77
K+L HLR HPEL G A +S +RR E+ A A GE A+ P
Sbjct 191 KSLARHLRLHPELSGPGVAAKVLAAS---VRRAKGPEE--AVAADGEIAI-------PVG 238
Query 78 DGRGRI 83
DG G I
Sbjct 239 DGEGII 244
> dre:100001432 eKir-like; K05009 potassium inwardly-rectifying
channel subfamily J member 16
Length=494
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 20/55 (36%), Positives = 30/55 (54%), Gaps = 1/55 (1%)
Query 32 GEDAALMT-TGSSLALLRRLAVLEQQGAEALFGEPALQLEDILQPANDGRGRIHL 85
GE ++++ GSS + L+ LE +G+ AL GE + LED A +G G L
Sbjct 12 GEGSSVLEGEGSSALEDKGLSALENEGSSALKGEGSSALEDEGSSALEGEGSSEL 66
> cpv:cgd3_270 hypothetical protein
Length=1130
Score = 28.9 bits (63), Expect = 4.6, Method: Composition-based stats.
Identities = 16/37 (43%), Positives = 19/37 (51%), Gaps = 0/37 (0%)
Query 1 FKVADREGLLLLDLKDLKALLNHLRYHPELLGEDAAL 37
FKV DR +L L K L L L Y+P +L D L
Sbjct 432 FKVQDRINILPLKFKSLNDKLYSLCYYPYILTSDICL 468
Lambda K H
0.321 0.141 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2030857360
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40