bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2487_orf1
Length=157
Score E
Sequences producing significant alignments: (Bits) Value
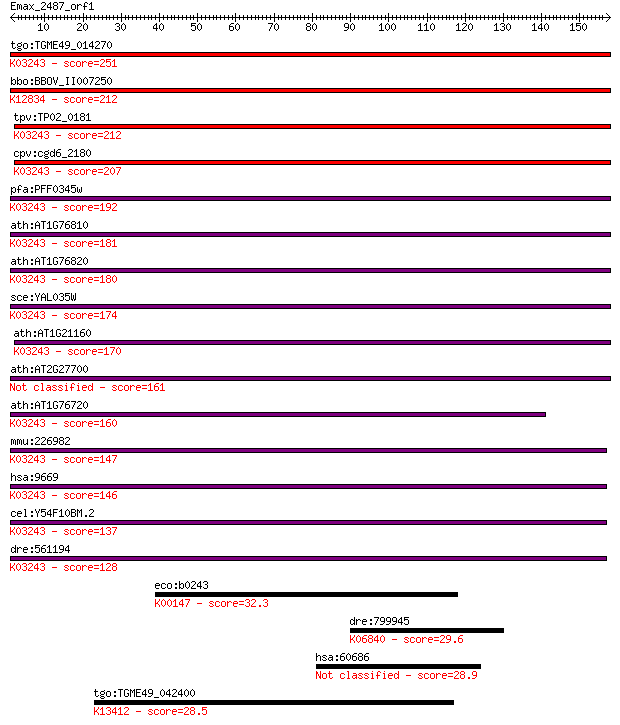
tgo:TGME49_014270 translation initiation factor IF-2, putative... 251 8e-67
bbo:BBOV_II007250 18.m06603; translation initiation factor IF-... 212 3e-55
tpv:TP02_0181 translation initiation factor IF-2; K03243 trans... 212 5e-55
cpv:cgd6_2180 Fun12p GTpase; translation initiation factor IF2... 207 8e-54
pfa:PFF0345w translation initiation factor IF-2, putative; K03... 192 4e-49
ath:AT1G76810 eukaryotic translation initiation factor 2 famil... 181 1e-45
ath:AT1G76820 GTP binding / GTPase; K03243 translation initiat... 180 2e-45
sce:YAL035W FUN12, yIF2; Fun12p; K03243 translation initiation... 174 7e-44
ath:AT1G21160 GTP binding / GTPase/ translation initiation fac... 170 2e-42
ath:AT2G27700 eukaryotic translation initiation factor 2 famil... 161 9e-40
ath:AT1G76720 GTP binding / GTPase/ translation initiation fac... 160 1e-39
mmu:226982 Eif5b, A030003E17Rik, BC018347, IF2; eukaryotic tra... 147 2e-35
hsa:9669 EIF5B, DKFZp434I036, FLJ10524, IF2, KIAA0741; eukaryo... 146 2e-35
cel:Y54F10BM.2 iffb-1; Initiation Factor Five B (eIF5B) family... 137 2e-32
dre:561194 im:6912504; si:ch211-254d18.3; K03243 translation i... 128 6e-30
eco:b0243 proA, ECK0244, JW0233, pro; gamma-glutamylphosphate ... 32.3 0.65
dre:799945 sema3fb; semaphorin 3fb; K06840 semaphorin 3 29.6
hsa:60686 C14orf93, FLJ12154; chromosome 14 open reading frame 93 28.9
tgo:TGME49_042400 calcium-dependent protein kinase, putative (... 28.5 8.8
> tgo:TGME49_014270 translation initiation factor IF-2, putative
(EC:2.7.7.4); K03243 translation initiation factor 5B
Length=1144
Score = 251 bits (640), Expect = 8e-67, Method: Compositional matrix adjust.
Identities = 119/157 (75%), Positives = 139/157 (88%), Gaps = 0/157 (0%)
Query 1 TTIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMG 60
TTIDVILV G LYEGDKIV+CG+ GPI+TTIR LLTPQPLKELRVKGEYI H+F++A+MG
Sbjct 797 TTIDVILVHGTLYEGDKIVVCGMSGPIVTTIRALLTPQPLKELRVKGEYIQHAFIQAAMG 856
Query 61 VKIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGS 120
VKI A L+EAVAGTS+FVVEE D I D++ EVM DMG++ KSVD TGSGV+V ASTLGS
Sbjct 857 VKISAPNLEEAVAGTSVFVVEEDDDIEDLKEEVMSDMGSIFKSVDRTGSGVYVMASTLGS 916
Query 121 LEALLVYLEECKIPVFAVSIGTVQKKDIKRASVMREK 157
LEALLV+L+E KIPVF V+IGTVQKKD+K+AS+MREK
Sbjct 917 LEALLVFLQESKIPVFGVNIGTVQKKDVKKASIMREK 953
> bbo:BBOV_II007250 18.m06603; translation initiation factor IF-2;
K12834 PHD finger-like domain-containing protein 5A
Length=1033
Score = 212 bits (540), Expect = 3e-55, Method: Compositional matrix adjust.
Identities = 102/157 (64%), Positives = 131/157 (83%), Gaps = 0/157 (0%)
Query 1 TTIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMG 60
TT+DVIL+ G + EGDKIV+CGL GPI+TTIRTLLTPQPL ELRVKGEY+ H+ ++A+MG
Sbjct 685 TTVDVILLSGTINEGDKIVLCGLSGPIVTTIRTLLTPQPLAELRVKGEYVKHTSIKAAMG 744
Query 61 VKIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGS 120
VK+VA GL+E VAGT L +VE+ D I + +VM+DM ++ +V+ TG GV+V ASTLGS
Sbjct 745 VKLVAQGLEETVAGTELLLVEDDDDIEQLCEDVMQDMSSIFGNVNRTGVGVYVMASTLGS 804
Query 121 LEALLVYLEECKIPVFAVSIGTVQKKDIKRASVMREK 157
LEALL +L + KIPVF+V+IGTVQKKD+K+AS+MREK
Sbjct 805 LEALLQFLTDKKIPVFSVNIGTVQKKDVKKASIMREK 841
> tpv:TP02_0181 translation initiation factor IF-2; K03243 translation
initiation factor 5B
Length=921
Score = 212 bits (539), Expect = 5e-55, Method: Compositional matrix adjust.
Identities = 100/156 (64%), Positives = 128/156 (82%), Gaps = 0/156 (0%)
Query 2 TIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMGV 61
T+DVIL+ G+L EGD+IV+CGL GPI+TTIRTLLTPQPL ELRVKGEY+ HS ++A+M V
Sbjct 574 TVDVILLDGILREGDRIVLCGLSGPIVTTIRTLLTPQPLSELRVKGEYVKHSHIKAAMSV 633
Query 62 KIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGSL 121
KIVA GLD+ VAGT LFVV EGD + ++ EVM D+ ++ +D TG GV+V ASTLGSL
Sbjct 634 KIVANGLDDTVAGTELFVVGEGDDVDELCTEVMSDISSIFDCIDRTGIGVYVMASTLGSL 693
Query 122 EALLVYLEECKIPVFAVSIGTVQKKDIKRASVMREK 157
EALL +L + KI +++V+IG VQKKD+K+AS+MREK
Sbjct 694 EALLHFLNDKKIKIYSVNIGPVQKKDVKKASIMREK 729
> cpv:cgd6_2180 Fun12p GTpase; translation initiation factor IF2
; K03243 translation initiation factor 5B
Length=896
Score = 207 bits (528), Expect = 8e-54, Method: Compositional matrix adjust.
Identities = 101/159 (63%), Positives = 124/159 (77%), Gaps = 3/159 (1%)
Query 2 TIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMGV 61
TIDVILV G+L EGD I++CGL PI+TTIR LLTPQP+ E+RVKGEYI H F++ASMGV
Sbjct 544 TIDVILVSGILREGDTIIVCGLSAPIVTTIRALLTPQPMHEMRVKGEYIHHRFIKASMGV 603
Query 62 KIVAAGLDEAVAGTSLFVVEEGDSISDIEG---EVMKDMGAVLKSVDSTGSGVFVAASTL 118
KI A GLD+AVAGT L V + + +IE EVMKDMG + SVD TG+GV+V ASTL
Sbjct 604 KICANGLDDAVAGTQLLVQSKNSTPEEIESLKEEVMKDMGDIFSSVDRTGNGVYVMASTL 663
Query 119 GSLEALLVYLEECKIPVFAVSIGTVQKKDIKRASVMREK 157
GSLEALLV+L+ IPV A++IGTV K D++RAS+M E+
Sbjct 664 GSLEALLVFLKSSNIPVVALNIGTVHKSDVRRASIMHER 702
> pfa:PFF0345w translation initiation factor IF-2, putative; K03243
translation initiation factor 5B
Length=977
Score = 192 bits (488), Expect = 4e-49, Method: Composition-based stats.
Identities = 94/157 (59%), Positives = 121/157 (77%), Gaps = 0/157 (0%)
Query 1 TTIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMG 60
TTIDVIL G+L E D IV+CG++GPI+T IR LLTPQPLKELR+K EYI H +++A +G
Sbjct 631 TTIDVILTNGILRESDTIVLCGINGPIVTVIRALLTPQPLKELRIKNEYIHHKYIKACIG 690
Query 61 VKIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGS 120
VKI A L+E + GTSLFVV + + + +VM D+ V VD TG G++V ASTLGS
Sbjct 691 VKISANNLEEVLCGTSLFVVNNIEEEEEYKKKVMTDVSDVFNHVDKTGVGLYVMASTLGS 750
Query 121 LEALLVYLEECKIPVFAVSIGTVQKKDIKRASVMREK 157
LEALL++L + KIPVF V+IGT+QKKD+K+AS+MREK
Sbjct 751 LEALLIFLNDSKIPVFGVNIGTIQKKDVKKASIMREK 787
> ath:AT1G76810 eukaryotic translation initiation factor 2 family
protein / eIF-2 family protein; K03243 translation initiation
factor 5B
Length=1294
Score = 181 bits (458), Expect = 1e-45, Method: Compositional matrix adjust.
Identities = 91/159 (57%), Positives = 117/159 (73%), Gaps = 2/159 (1%)
Query 1 TTIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMG 60
TTIDV+LV G L+EGD+IV+CGL GPI+TTIR LLTP P+KELRVKG Y+ + ++A+ G
Sbjct 943 TTIDVVLVNGELHEGDQIVVCGLQGPIVTTIRALLTPHPMKELRVKGTYLHYKEIKAAQG 1002
Query 61 VKIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGS 120
+KI A GL+ A+AGT+L VV D I I+ M+DM +VL +D +G GV+V ASTLGS
Sbjct 1003 IKITAQGLEHAIAGTALHVVGPDDDIEAIKESAMEDMESVLSRIDKSGEGVYVQASTLGS 1062
Query 121 LEALLVYLEE--CKIPVFAVSIGTVQKKDIKRASVMREK 157
LEALL YL+ KIPV + IG V KKD+ +A VM E+
Sbjct 1063 LEALLEYLKSPAVKIPVSGIGIGPVHKKDVMKAGVMLER 1101
> ath:AT1G76820 GTP binding / GTPase; K03243 translation initiation
factor 5B
Length=1166
Score = 180 bits (456), Expect = 2e-45, Method: Compositional matrix adjust.
Identities = 91/159 (57%), Positives = 115/159 (72%), Gaps = 2/159 (1%)
Query 1 TTIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMG 60
TTIDV+LV G L+EGD+IV+CGL GPI+TTIR LLTP P+KELRVKG Y+ H ++A+ G
Sbjct 815 TTIDVVLVNGELHEGDQIVVCGLQGPIVTTIRALLTPHPMKELRVKGTYLHHKEIKAAQG 874
Query 61 VKIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGS 120
+KI A GL+ A+AGTSL VV D I ++ M+DM +VL +D +G GV+V STLGS
Sbjct 875 IKITAQGLEHAIAGTSLHVVGPDDDIEAMKESAMEDMESVLSRIDKSGEGVYVQTSTLGS 934
Query 121 LEALLVYLE--ECKIPVFAVSIGTVQKKDIKRASVMREK 157
LEALL +L+ IPV + IG V KKDI +A VM EK
Sbjct 935 LEALLEFLKTPAVNIPVSGIGIGPVHKKDIMKAGVMLEK 973
> sce:YAL035W FUN12, yIF2; Fun12p; K03243 translation initiation
factor 5B
Length=1002
Score = 174 bits (442), Expect = 7e-44, Method: Compositional matrix adjust.
Identities = 88/157 (56%), Positives = 118/157 (75%), Gaps = 0/157 (0%)
Query 1 TTIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMG 60
TTIDVIL G L EGD+IV+CG++GPI+T IR LLTPQPL+ELR+K EY+ H V+A++G
Sbjct 645 TTIDVILSNGYLREGDRIVLCGMNGPIVTNIRALLTPQPLRELRLKSEYVHHKEVKAALG 704
Query 61 VKIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGS 120
VKI A L++AV+G+ L VV D ++ +VM D+ +L SVD+TG GV V ASTLGS
Sbjct 705 VKIAANDLEKAVSGSRLLVVGPEDDEDELMDDVMDDLTGLLDSVDTTGKGVVVQASTLGS 764
Query 121 LEALLVYLEECKIPVFAVSIGTVQKKDIKRASVMREK 157
LEALL +L++ KIPV ++ +G V K+D+ +AS M EK
Sbjct 765 LEALLDFLKDMKIPVMSIGLGPVYKRDVMKASTMLEK 801
> ath:AT1G21160 GTP binding / GTPase/ translation initiation factor;
K03243 translation initiation factor 5B
Length=1092
Score = 170 bits (430), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 83/158 (52%), Positives = 115/158 (72%), Gaps = 2/158 (1%)
Query 2 TIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMGV 61
T+DV+LV GVL EGD+IV+CG GPI+TTIR+LLTP P+ E+RV G Y+ H V+A+ G+
Sbjct 736 TVDVVLVNGVLREGDQIVVCGSQGPIVTTIRSLLTPYPMNEMRVTGTYMPHREVKAAQGI 795
Query 62 KIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGSL 121
KI A GL+ A+AGT+L V+ + + + + M+D+ +V+ +D +G GV+V ASTLGSL
Sbjct 796 KIAAQGLEHAIAGTALHVIGPNEDMEEAKKNAMEDIESVMNRIDKSGEGVYVQASTLGSL 855
Query 122 EALLVYLE--ECKIPVFAVSIGTVQKKDIKRASVMREK 157
EALL +L+ + KIPV + IG V KKDI +A VM EK
Sbjct 856 EALLEFLKSSDVKIPVSGIGIGPVHKKDIMKAGVMLEK 893
> ath:AT2G27700 eukaryotic translation initiation factor 2 family
protein / eIF-2 family protein
Length=480
Score = 161 bits (407), Expect = 9e-40, Method: Compositional matrix adjust.
Identities = 81/159 (50%), Positives = 112/159 (70%), Gaps = 2/159 (1%)
Query 1 TTIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMG 60
TTIDV+LV G L+EG +IV+CGL GPI+TTIR LLTP P+KEL V G ++ H ++A+
Sbjct 275 TTIDVVLVNGELHEGGQIVVCGLQGPIVTTIRALLTPHPIKELHVNGNHVHHEVIKAAEC 334
Query 61 VKIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGS 120
+ I+A L+ + GT+L VV D I I+ VM+D+ +VL +D +G GV++ ASTLGS
Sbjct 335 INIIAKDLEHVIVGTALHVVGPDDDIEAIKELVMEDVNSVLSRIDKSGEGVYIQASTLGS 394
Query 121 LEALLVYLEE--CKIPVFAVSIGTVQKKDIKRASVMREK 157
LEALL +L+ K+PV + IG VQKKD+ +A VM E+
Sbjct 395 LEALLEFLKSPAVKLPVGGIGIGPVQKKDVMKAGVMLER 433
> ath:AT1G76720 GTP binding / GTPase/ translation initiation factor;
K03243 translation initiation factor 5B
Length=1191
Score = 160 bits (406), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 78/140 (55%), Positives = 101/140 (72%), Gaps = 3/140 (2%)
Query 1 TTIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMG 60
TTIDV+LV G L+EGD+IV+CGL GPI+TTIR LLTP P+KELRVKG Y+ H ++A+ G
Sbjct 866 TTIDVVLVNGELHEGDQIVVCGLQGPIVTTIRALLTPHPMKELRVKGTYLHHKEIKAAQG 925
Query 61 VKIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGS 120
+KI A GL+ A+AGTSL VV D I ++ M+DM +VL +D +G GV+V STLGS
Sbjct 926 IKITAQGLEHAIAGTSLHVVGPDDDIEAMKESAMEDMESVLSRIDKSGEGVYVQTSTLGS 985
Query 121 LEALLVYLEECKIPVFAVSI 140
LEALL +L K P + +
Sbjct 986 LEALLEFL---KTPAVNIPV 1002
> mmu:226982 Eif5b, A030003E17Rik, BC018347, IF2; eukaryotic translation
initiation factor 5B; K03243 translation initiation
factor 5B
Length=1216
Score = 147 bits (370), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 75/156 (48%), Positives = 105/156 (67%), Gaps = 0/156 (0%)
Query 1 TTIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMG 60
TTIDVIL+ G L EGD I++ G+ GPI+T IR LL P P+KELRVK +Y H VEA+ G
Sbjct 867 TTIDVILINGRLKEGDTIIVPGVEGPIVTQIRGLLLPPPMKELRVKNQYEKHKEVEAAQG 926
Query 61 VKIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGS 120
VKI+ L++ +AG L V + D I ++ E++ ++ L ++ GV+V ASTLGS
Sbjct 927 VKILGKDLEKTLAGLPLLVAYKDDEIPVLKDELIHELKQTLNAIKLEEKGVYVQASTLGS 986
Query 121 LEALLVYLEECKIPVFAVSIGTVQKKDIKRASVMRE 156
LEALL +L+ ++P ++IG V KKD+ +ASVM E
Sbjct 987 LEALLEFLKTSEVPYAGINIGPVHKKDVMKASVMLE 1022
> hsa:9669 EIF5B, DKFZp434I036, FLJ10524, IF2, KIAA0741; eukaryotic
translation initiation factor 5B; K03243 translation initiation
factor 5B
Length=1220
Score = 146 bits (369), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 75/156 (48%), Positives = 105/156 (67%), Gaps = 0/156 (0%)
Query 1 TTIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMG 60
TTIDVIL+ G L EGD I++ G+ GPI+T IR LL P P+KELRVK +Y H VEA+ G
Sbjct 871 TTIDVILINGRLKEGDTIIVPGVEGPIVTQIRGLLLPPPMKELRVKNQYEKHKEVEAAQG 930
Query 61 VKIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGS 120
VKI+ L++ +AG L V + D I ++ E++ ++ L ++ GV+V ASTLGS
Sbjct 931 VKILGKDLEKTLAGLPLLVAYKEDEIPVLKDELIHELKQTLNAIKLEEKGVYVQASTLGS 990
Query 121 LEALLVYLEECKIPVFAVSIGTVQKKDIKRASVMRE 156
LEALL +L+ ++P ++IG V KKD+ +ASVM E
Sbjct 991 LEALLEFLKTSEVPYAGINIGPVHKKDVMKASVMLE 1026
> cel:Y54F10BM.2 iffb-1; Initiation Factor Five B (eIF5B) family
member (iffb-1); K03243 translation initiation factor 5B
Length=1074
Score = 137 bits (344), Expect = 2e-32, Method: Composition-based stats.
Identities = 71/156 (45%), Positives = 101/156 (64%), Gaps = 0/156 (0%)
Query 1 TTIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMG 60
TTIDVILV G + GD IV+ G G I T +R LL P+PLKELRVK EYI + V+ + G
Sbjct 725 TTIDVILVNGTMRAGDVIVLTGSDGAITTQVRELLMPKPLKELRVKNEYIHYKEVKGARG 784
Query 61 VKIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGS 120
VK++A L++ +AG +++ + D + + E + + L ++ GV+V ASTLGS
Sbjct 785 VKVLAKNLEKVLAGLPIYITDREDEVDYLRHEADRQLANALHAIRKKPEGVYVQASTLGS 844
Query 121 LEALLVYLEECKIPVFAVSIGTVQKKDIKRASVMRE 156
LEALL +L+ IP V+IG V KKD+++AS M+E
Sbjct 845 LEALLEFLKSQNIPYSNVNIGPVHKKDVQKASAMKE 880
> dre:561194 im:6912504; si:ch211-254d18.3; K03243 translation
initiation factor 5B
Length=1166
Score = 128 bits (322), Expect = 6e-30, Method: Compositional matrix adjust.
Identities = 73/156 (46%), Positives = 101/156 (64%), Gaps = 0/156 (0%)
Query 1 TTIDVILVQGVLYEGDKIVICGLHGPIITTIRTLLTPQPLKELRVKGEYISHSFVEASMG 60
TTIDVIL+ G L EGD I++ G+ GPI+T IR LL P PLKELRVK +Y H V + G
Sbjct 817 TTIDVILINGHLREGDTIIVPGVDGPIVTQIRGLLLPPPLKELRVKNQYEKHKEVCTAQG 876
Query 61 VKIVAAGLDEAVAGTSLFVVEEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGS 120
VKI+ L++ +AG L V + D + + E+++++ L ++ GV+V ASTLGS
Sbjct 877 VKILGKDLEKTLAGLPLLVAHKEDEVPVLRDELVRELKQTLNAIKLEEKGVYVQASTLGS 936
Query 121 LEALLVYLEECKIPVFAVSIGTVQKKDIKRASVMRE 156
LEALL +L K+P ++IG V KKD+ +AS M E
Sbjct 937 LEALLEFLRTSKVPYAGINIGPVHKKDVMKASTMLE 972
> eco:b0243 proA, ECK0244, JW0233, pro; gamma-glutamylphosphate
reductase (EC:1.2.1.41); K00147 glutamate-5-semialdehyde dehydrogenase
[EC:1.2.1.41]
Length=417
Score = 32.3 bits (72), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 40/80 (50%), Gaps = 8/80 (10%)
Query 39 PLKELRVKGEYISHSFVEASMGVKIVAAGLDEAVAGTSLFVVEEGDSISD-IEGEVMKDM 97
P K + VK E F+ + VKIV + LD+A+A + E G SD I M++
Sbjct 298 PAKVVAVKAEEYDDEFLSLDLNVKIV-SDLDDAIA----HIREHGTQHSDAILTRDMRNA 352
Query 98 GAVLKSVDSTGSGVFVAAST 117
+ VDS S V+V AST
Sbjct 353 QRFVNEVDS--SAVYVNAST 370
> dre:799945 sema3fb; semaphorin 3fb; K06840 semaphorin 3
Length=758
Score = 29.6 bits (65), Expect = 4.2, Method: Composition-based stats.
Identities = 14/41 (34%), Positives = 27/41 (65%), Gaps = 1/41 (2%)
Query 90 EGEVMK-DMGAVLKSVDSTGSGVFVAASTLGSLEALLVYLE 129
+G V+K D G +L+S+ S+ SG+++ +T + + LV L+
Sbjct 623 DGRVLKTDQGLLLRSLQSSDSGIYICTATEKNFKHTLVKLQ 663
> hsa:60686 C14orf93, FLJ12154; chromosome 14 open reading frame
93
Length=538
Score = 28.9 bits (63), Expect = 6.8, Method: Composition-based stats.
Identities = 17/43 (39%), Positives = 21/43 (48%), Gaps = 0/43 (0%)
Query 81 EEGDSISDIEGEVMKDMGAVLKSVDSTGSGVFVAASTLGSLEA 123
EE + + GE K + AV + DS GSGV V L L A
Sbjct 122 EEPGPLKESPGEAFKALSAVEEECDSVGSGVQVVIEELRQLGA 164
> tgo:TGME49_042400 calcium-dependent protein kinase, putative
(EC:2.7.11.17); K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=604
Score = 28.5 bits (62), Expect = 8.8, Method: Composition-based stats.
Identities = 31/102 (30%), Positives = 51/102 (50%), Gaps = 12/102 (11%)
Query 23 LHGPIITTIRTLLTPQPLKE--LRVKGEYISHSFVEASMGVKIVAAGLDEAVAGT-SLFV 79
L+ I+ R LK+ L V + ++ ++A +K + LDE GT ++
Sbjct 398 LNSKILDNFRAFRAVSKLKKAALTVIAQQMNEGQIKA---LKNIFLALDEDGDGTLTINE 454
Query 80 VEEGDSISDIEGEVMKDMGAVLKSVDSTGSGV-----FVAAS 116
+ G S S ++ E+ D+ A++ VDS GSGV F+AAS
Sbjct 455 IRVGLSKSGLK-EMPSDLDALMNEVDSDGSGVIDYTEFIAAS 495
Lambda K H
0.317 0.136 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3516928200
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40