bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2476_orf1
Length=110
Score E
Sequences producing significant alignments: (Bits) Value
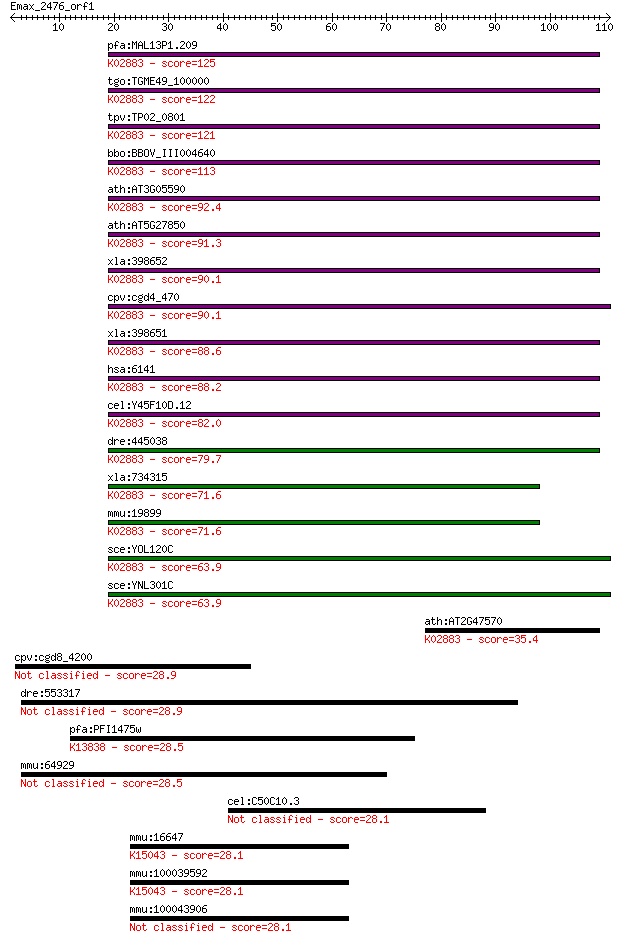
pfa:MAL13P1.209 60S ribosomal protein L18-2, putative; K02883 ... 125 3e-29
tgo:TGME49_100000 60S ribosomal protein L18, putative ; K02883... 122 2e-28
tpv:TP02_0801 60S ribosomal protein L18; K02883 large subunit ... 121 5e-28
bbo:BBOV_III004640 17.m07415; 60S ribosomal protein L18; K0288... 113 1e-25
ath:AT3G05590 RPL18; RPL18 (RIBOSOMAL PROTEIN L18); structural... 92.4 3e-19
ath:AT5G27850 60S ribosomal protein L18 (RPL18C); K02883 large... 91.3 6e-19
xla:398652 rpl18-b, L14B, MGC130777, MGC64299, rpl14b; ribosom... 90.1 1e-18
cpv:cgd4_470 60S ribosomal protein L18 ; K02883 large subunit ... 90.1 2e-18
xla:398651 rpl18-a, MGC64315, rpl14a, rpl18; ribosomal protein... 88.6 4e-18
hsa:6141 RPL18; ribosomal protein L18; K02883 large subunit ri... 88.2 6e-18
cel:Y45F10D.12 rpl-18; Ribosomal Protein, Large subunit family... 82.0 4e-16
dre:445038 fa98d03, wu:fa98d03; zgc:92872; K02883 large subuni... 79.7 2e-15
xla:734315 hypothetical protein MGC98504; K02883 large subunit... 71.6 5e-13
mmu:19899 Rpl18, L18, Rpl18a; ribosomal protein L18; K02883 la... 71.6 5e-13
sce:YOL120C RPL18A, RP28A; Rpl18ap; K02883 large subunit ribos... 63.9 1e-10
sce:YNL301C RPL18B, RP28B; Rpl18bp; K02883 large subunit ribos... 63.9 1e-10
ath:AT2G47570 60S ribosomal protein L18 (RPL18A); K02883 large... 35.4 0.045
cpv:cgd8_4200 hypothetical protein 28.9 4.1
dre:553317 ZNF261; si:ch211-173p18.3 28.9 4.4
pfa:PFI1475w MSP1, MSA1, MSP-1, Pf190, PMMSA; merozoite surfac... 28.5 5.4
mmu:64929 Scel, 9230114I02Rik, MGC132819, MGC132820; sciellin 28.5 5.9
cel:C50C10.3 sru-30; Serpentine Receptor, class U family membe... 28.1 6.4
mmu:16647 Kpna2, 2410044B12Rik, IPOA1, MGC91246, Rch1; karyoph... 28.1 6.7
mmu:100039592 Gm10259; predicted pseudogene 10259; K15043 impo... 28.1 6.7
mmu:100043906 Gm10184; predicted pseudogene 10184 28.1
> pfa:MAL13P1.209 60S ribosomal protein L18-2, putative; K02883
large subunit ribosomal protein L18e
Length=187
Score = 125 bits (315), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 63/90 (70%), Positives = 69/90 (76%), Gaps = 0/90 (0%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MGI LKN GR KKHGR+ LVS+NPYLRL VKLY FLARRTN+NFNK+I KRL+ P+R R
Sbjct 1 MGIALKNVGRIKKHGRKHLVSKNPYLRLLVKLYNFLARRTNANFNKIIAKRLIMPKRYRP 60
Query 79 PLSLSKLSMRMRKEENKTAVVVGSIVDDPR 108
PLSLSKL M N AVVVGSI DD R
Sbjct 61 PLSLSKLQYHMANHPNDIAVVVGSITDDKR 90
> tgo:TGME49_100000 60S ribosomal protein L18, putative ; K02883
large subunit ribosomal protein L18e
Length=187
Score = 122 bits (307), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 60/90 (66%), Positives = 73/90 (81%), Gaps = 0/90 (0%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MGIDL+ GR KK GRR + S+NPYLRL VKLYQFLARRTNS FN V+LKRLM P+RL+A
Sbjct 1 MGIDLERNGRKKKGGRRTVKSQNPYLRLLVKLYQFLARRTNSRFNAVVLKRLMNPKRLKA 60
Query 79 PLSLSKLSMRMRKEENKTAVVVGSIVDDPR 108
P+SLSKL+ M+ +E+ AV+VGS+ DD R
Sbjct 61 PMSLSKLARHMKGKEDHIAVLVGSVTDDIR 90
> tpv:TP02_0801 60S ribosomal protein L18; K02883 large subunit
ribosomal protein L18e
Length=184
Score = 121 bits (304), Expect = 5e-28, Method: Compositional matrix adjust.
Identities = 60/90 (66%), Positives = 73/90 (81%), Gaps = 0/90 (0%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MGIDL GR KK GR+ALVS+NPYLRL VKLY+FL+RRT+ +FNKV+LKRL+ PRR +A
Sbjct 1 MGIDLVKAGRVKKPGRKALVSKNPYLRLLVKLYKFLSRRTDHSFNKVVLKRLLMPRRFKA 60
Query 79 PLSLSKLSMRMRKEENKTAVVVGSIVDDPR 108
PLSLSKLS M K + TAVVVG++ +D R
Sbjct 61 PLSLSKLSKHMEKRPDNTAVVVGTVTNDER 90
> bbo:BBOV_III004640 17.m07415; 60S ribosomal protein L18; K02883
large subunit ribosomal protein L18e
Length=187
Score = 113 bits (283), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 54/90 (60%), Positives = 69/90 (76%), Gaps = 0/90 (0%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MGIDL+ GR KK GR+ALVS++PYL+L V Y+ LARRT S FN+ ILKRL+ PRR +
Sbjct 1 MGIDLEKGGRVKKPGRKALVSQDPYLQLLVNSYKLLARRTQSKFNRTILKRLIMPRRFKC 60
Query 79 PLSLSKLSMRMRKEENKTAVVVGSIVDDPR 108
P+SLSKL+ M+ E+ TAV+VG+I DD R
Sbjct 61 PMSLSKLTKHMKGREHTTAVIVGTITDDIR 90
> ath:AT3G05590 RPL18; RPL18 (RIBOSOMAL PROTEIN L18); structural
constituent of ribosome; K02883 large subunit ribosomal protein
L18e
Length=187
Score = 92.4 bits (228), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 51/90 (56%), Positives = 64/90 (71%), Gaps = 0/90 (0%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MGIDL G++KK R A S++ YL+L VKLY+FL RRTNS FN VILKRL + +A
Sbjct 1 MGIDLIAGGKSKKTKRTAPKSDDVYLKLTVKLYRFLVRRTNSKFNGVILKRLFMSKVNKA 60
Query 79 PLSLSKLSMRMRKEENKTAVVVGSIVDDPR 108
PLSLS+L M +E+K AV+VG+I DD R
Sbjct 61 PLSLSRLVEFMTGKEDKIAVLVGTITDDLR 90
> ath:AT5G27850 60S ribosomal protein L18 (RPL18C); K02883 large
subunit ribosomal protein L18e
Length=187
Score = 91.3 bits (225), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 50/90 (55%), Positives = 65/90 (72%), Gaps = 0/90 (0%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MGIDL G++KK R A S++ YL+L VKLY+FL RR+NSNFN VILKRL + +A
Sbjct 1 MGIDLIAGGKSKKTKRTAPKSDDVYLKLLVKLYRFLVRRSNSNFNAVILKRLFMSKVNKA 60
Query 79 PLSLSKLSMRMRKEENKTAVVVGSIVDDPR 108
PLSLS+L M +++K AV+VG+I DD R
Sbjct 61 PLSLSRLVEFMTGKDDKIAVLVGTITDDLR 90
> xla:398652 rpl18-b, L14B, MGC130777, MGC64299, rpl14b; ribosomal
protein L18; K02883 large subunit ribosomal protein L18e
Length=188
Score = 90.1 bits (222), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 51/92 (55%), Positives = 68/92 (73%), Gaps = 3/92 (3%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MGID+++ + +K R+ S++ YLRL VKLY+FLARRTNS+FN+V+LKRL R R
Sbjct 1 MGIDIRH-NKDRKVRRKEPKSQDIYLRLLVKLYRFLARRTNSSFNRVVLKRLFMSRTNRP 59
Query 79 PLSLSKL--SMRMRKEENKTAVVVGSIVDDPR 108
PLS+S+L M+++ ENKTAVVVG I DD R
Sbjct 60 PLSMSRLIRKMKLQGRENKTAVVVGCITDDVR 91
> cpv:cgd4_470 60S ribosomal protein L18 ; K02883 large subunit
ribosomal protein L18e
Length=190
Score = 90.1 bits (222), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 47/92 (51%), Positives = 58/92 (63%), Gaps = 0/92 (0%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MGIDLK GR KK + S NPY L KLY FL RRT+S+FN +L RL RR +A
Sbjct 4 MGIDLKAGGRVKKTQSKRTRSTNPYRILLSKLYTFLGRRTDSSFNDTVLHRLKMARRFQA 63
Query 79 PLSLSKLSMRMRKEENKTAVVVGSIVDDPRTL 110
P+S+S+L M +E K A VVG++ DD R L
Sbjct 64 PISVSRLVQFMNGKEGKIAAVVGTVTDDTRVL 95
> xla:398651 rpl18-a, MGC64315, rpl14a, rpl18; ribosomal protein
L18; K02883 large subunit ribosomal protein L18e
Length=188
Score = 88.6 bits (218), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 51/92 (55%), Positives = 67/92 (72%), Gaps = 3/92 (3%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MGID+++ + +K R+ S++ YLRL VKLY+FLARRTNS+FN+V+LKRL R R
Sbjct 1 MGIDIRH-NKDRKVRRKEPKSQDIYLRLLVKLYRFLARRTNSSFNRVVLKRLFMSRTNRP 59
Query 79 PLSLSKL--SMRMRKEENKTAVVVGSIVDDPR 108
PLS+S+L M++ ENKTAVVVG I DD R
Sbjct 60 PLSMSRLIRKMKLPGRENKTAVVVGCITDDVR 91
> hsa:6141 RPL18; ribosomal protein L18; K02883 large subunit
ribosomal protein L18e
Length=188
Score = 88.2 bits (217), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 50/92 (54%), Positives = 67/92 (72%), Gaps = 3/92 (3%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MG+D+++ + +K R+ S++ YLRL VKLY+FLARRTNS FN+V+LKRL R R
Sbjct 1 MGVDIRH-NKDRKVRRKEPKSQDIYLRLLVKLYRFLARRTNSTFNQVVLKRLFMSRTNRP 59
Query 79 PLSLSKL--SMRMRKEENKTAVVVGSIVDDPR 108
PLSLS++ M++ ENKTAVVVG+I DD R
Sbjct 60 PLSLSRMIRKMKLPGRENKTAVVVGTITDDVR 91
> cel:Y45F10D.12 rpl-18; Ribosomal Protein, Large subunit family
member (rpl-18); K02883 large subunit ribosomal protein L18e
Length=188
Score = 82.0 bits (201), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 48/92 (52%), Positives = 59/92 (64%), Gaps = 3/92 (3%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MGID+ N + R A SENPYLRL KLY FLARRT FN ++LKRL RR R
Sbjct 1 MGIDI-NHKHDRVARRTAPKSENPYLRLLSKLYAFLARRTGEKFNAIVLKRLRMSRRNRQ 59
Query 79 PLSLSKLSMRMRK--EENKTAVVVGSIVDDPR 108
PLSL+KL+ ++K ENKT V + ++ DD R
Sbjct 60 PLSLAKLARAVQKAGNENKTVVTLSTVTDDAR 91
> dre:445038 fa98d03, wu:fa98d03; zgc:92872; K02883 large subunit
ribosomal protein L18e
Length=182
Score = 79.7 bits (195), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 46/92 (50%), Positives = 65/92 (70%), Gaps = 3/92 (3%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MG+D+++ K H R+ S++ YLRL VKLY+FL+RR+++ FNKVIL+RL + R
Sbjct 1 MGVDIRHNKDRKVH-RKEPKSQDIYLRLLVKLYRFLSRRSDAPFNKVILRRLFMSKTNRP 59
Query 79 PLSLSKL--SMRMRKEENKTAVVVGSIVDDPR 108
PL+LS+L M++ EN TAVVVG+I DD R
Sbjct 60 PLALSRLIRKMKLPGRENLTAVVVGTITDDVR 91
> xla:734315 hypothetical protein MGC98504; K02883 large subunit
ribosomal protein L18e
Length=188
Score = 71.6 bits (174), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 42/81 (51%), Positives = 58/81 (71%), Gaps = 3/81 (3%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MG+D+++ + +K R+ S++ YLRL VKLY+FLARRTNS FN+V+LKRL R R
Sbjct 1 MGVDIRH-NKDRKVRRKEPKSQDIYLRLLVKLYRFLARRTNSTFNQVVLKRLFMSRTNRP 59
Query 79 PLSLSKL--SMRMRKEENKTA 97
PLSLS++ M++ ENKTA
Sbjct 60 PLSLSRMIRKMKLPGRENKTA 80
> mmu:19899 Rpl18, L18, Rpl18a; ribosomal protein L18; K02883
large subunit ribosomal protein L18e
Length=188
Score = 71.6 bits (174), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 42/81 (51%), Positives = 58/81 (71%), Gaps = 3/81 (3%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MG+D+++ + +K R+ S++ YLRL VKLY+FLARRTNS FN+V+LKRL R R
Sbjct 1 MGVDIRH-NKDRKVRRKEPKSQDIYLRLLVKLYRFLARRTNSTFNQVVLKRLFMSRTNRP 59
Query 79 PLSLSKL--SMRMRKEENKTA 97
PLSLS++ M++ ENKTA
Sbjct 60 PLSLSRMIRKMKLPGRENKTA 80
> sce:YOL120C RPL18A, RP28A; Rpl18ap; K02883 large subunit ribosomal
protein L18e
Length=186
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 43/94 (45%), Positives = 62/94 (65%), Gaps = 2/94 (2%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MGID ++ + R A S+N YL+L VKLY FLARRT++ FNKV+LK L + R
Sbjct 1 MGIDHTSKQHKRSGHRTAPKSDNVYLKLLVKLYTFLARRTDAPFNKVVLKALFLSKINRP 60
Query 79 PLSLSKLSMRMRKE--ENKTAVVVGSIVDDPRTL 110
P+S+S+++ +++E NKT VVVG++ DD R
Sbjct 61 PVSVSRIARALKQEGAANKTVVVVGTVTDDARIF 94
> sce:YNL301C RPL18B, RP28B; Rpl18bp; K02883 large subunit ribosomal
protein L18e
Length=186
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 43/94 (45%), Positives = 62/94 (65%), Gaps = 2/94 (2%)
Query 19 MGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRA 78
MGID ++ + R A S+N YL+L VKLY FLARRT++ FNKV+LK L + R
Sbjct 1 MGIDHTSKQHKRSGHRTAPKSDNVYLKLLVKLYTFLARRTDAPFNKVVLKALFLSKINRP 60
Query 79 PLSLSKLSMRMRKE--ENKTAVVVGSIVDDPRTL 110
P+S+S+++ +++E NKT VVVG++ DD R
Sbjct 61 PVSVSRIARALKQEGAANKTVVVVGTVTDDARIF 94
> ath:AT2G47570 60S ribosomal protein L18 (RPL18A); K02883 large
subunit ribosomal protein L18e
Length=135
Score = 35.4 bits (80), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 16/32 (50%), Positives = 23/32 (71%), Gaps = 0/32 (0%)
Query 77 RAPLSLSKLSMRMRKEENKTAVVVGSIVDDPR 108
+APLSLS+L M ++ K AV+VG++ DD R
Sbjct 6 KAPLSLSRLVRYMDGKDGKIAVIVGTVTDDVR 37
> cpv:cgd8_4200 hypothetical protein
Length=913
Score = 28.9 bits (63), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 27/43 (62%), Gaps = 4/43 (9%)
Query 2 VFKPEKKNESLQEIKAKMGIDLKNRGRTKKHGRRALVSENPYL 44
+++ EK+NE LQE K ++ + RGR+ + G R ++ +N L
Sbjct 103 IYQLEKRNEELQEEKTRLEL----RGRSLEEGSREVLKKNEEL 141
> dre:553317 ZNF261; si:ch211-173p18.3
Length=1084
Score = 28.9 bits (63), Expect = 4.4, Method: Composition-based stats.
Identities = 25/91 (27%), Positives = 39/91 (42%), Gaps = 4/91 (4%)
Query 3 FKPEKKNESLQEIKAKMGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNF 62
F P K++ S + AK R R K G+ S+NP L V+LY+F + +
Sbjct 961 FYPPKEDSSDDGVPAKKRKKDDERQRVLKIGQN---SDNP-LHCPVRLYEFYLSKCSPGI 1016
Query 63 NKVILKRLMAPRRLRAPLSLSKLSMRMRKEE 93
K ++P R P S + S+ +E
Sbjct 1017 RHHATKFYLSPERSCVPSSPTWFSISALSDE 1047
> pfa:PFI1475w MSP1, MSA1, MSP-1, Pf190, PMMSA; merozoite surface
protein 1 precursor; K13838 merozoite surface protein 1
Length=1720
Score = 28.5 bits (62), Expect = 5.4, Method: Composition-based stats.
Identities = 19/67 (28%), Positives = 35/67 (52%), Gaps = 7/67 (10%)
Query 12 LQEIKAKMGIDLKNRGRTKKHGRRAL----VSENPYLRLAVKLYQFLARRTNSNFNKVIL 67
+ IK K+ ++ KN TK+ ++ L S+ Y L K Y+ + N+NFNK ++
Sbjct 474 INNIKKKIDLEEKNINHTKEQNKKLLEDYEKSKKDYEELLEKFYEM---KFNNNFNKDVV 530
Query 68 KRLMAPR 74
++ + R
Sbjct 531 DKIFSAR 537
> mmu:64929 Scel, 9230114I02Rik, MGC132819, MGC132820; sciellin
Length=652
Score = 28.5 bits (62), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 16/67 (23%), Positives = 35/67 (52%), Gaps = 6/67 (8%)
Query 3 FKPEKKNESLQEIKAKMGIDLKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNF 62
F KK+ + ++K+ G +L+ +G + RRA + +N +++ + Q + N+
Sbjct 4 FSSRKKSPTGNDLKSARGAELRQQGFQDVNKRRAFLQDNSWIKKPPEEEQ------DGNY 57
Query 63 NKVILKR 69
+V+L R
Sbjct 58 GRVVLNR 64
> cel:C50C10.3 sru-30; Serpentine Receptor, class U family member
(sru-30)
Length=340
Score = 28.1 bits (61), Expect = 6.4, Method: Composition-based stats.
Identities = 11/47 (23%), Positives = 27/47 (57%), Gaps = 0/47 (0%)
Query 41 NPYLRLAVKLYQFLARRTNSNFNKVILKRLMAPRRLRAPLSLSKLSM 87
+P+L ++L L+ + + +++++R + P + P SLS +S+
Sbjct 128 SPFLASLIRLVMILSPHNHGKYCRLLMRRFVFPFIIIVPFSLSLISL 174
> mmu:16647 Kpna2, 2410044B12Rik, IPOA1, MGC91246, Rch1; karyopherin
(importin) alpha 2; K15043 importin subunit alpha-2
Length=529
Score = 28.1 bits (61), Expect = 6.7, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 20/40 (50%), Gaps = 0/40 (0%)
Query 23 LKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNF 62
KN+G+ RR + N LR A K Q L RR S+F
Sbjct 17 FKNKGKDSTEMRRRRIEVNVELRKAKKDEQMLKRRNVSSF 56
> mmu:100039592 Gm10259; predicted pseudogene 10259; K15043 importin
subunit alpha-2
Length=529
Score = 28.1 bits (61), Expect = 6.7, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 20/40 (50%), Gaps = 0/40 (0%)
Query 23 LKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNF 62
KN+G+ RR + N LR A K Q L RR S+F
Sbjct 17 FKNKGKDSTEMRRRRIEVNVELRKAKKDEQMLKRRNVSSF 56
> mmu:100043906 Gm10184; predicted pseudogene 10184
Length=529
Score = 28.1 bits (61), Expect = 6.7, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 20/40 (50%), Gaps = 0/40 (0%)
Query 23 LKNRGRTKKHGRRALVSENPYLRLAVKLYQFLARRTNSNF 62
KN+G+ RR + N LR A K Q L RR S+F
Sbjct 17 FKNKGKDSTEMRRRRIEVNVELRKAKKDEQMLKRRNVSSF 56
Lambda K H
0.319 0.134 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2067351240
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40