bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
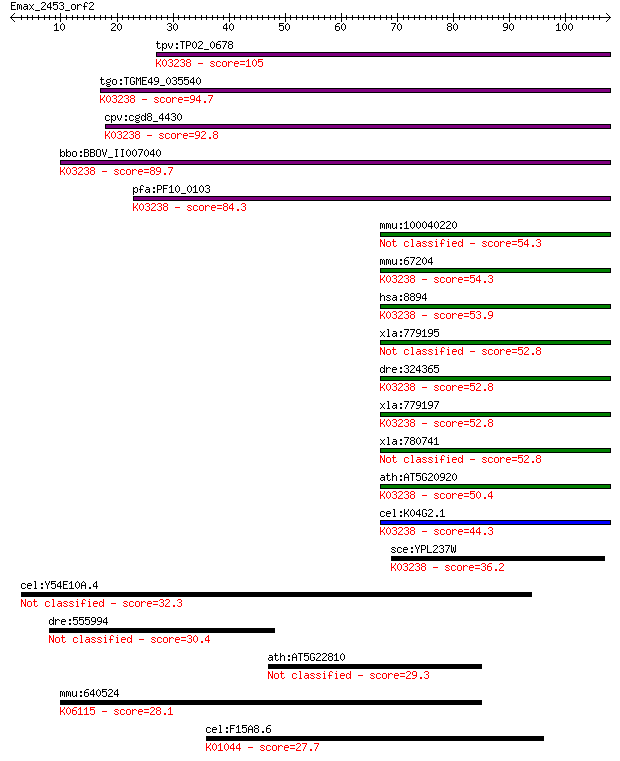
Query= Emax_2453_orf2
Length=107
Score E
Sequences producing significant alignments: (Bits) Value
tpv:TP02_0678 eukaryotic translation initiation factor 2 subun... 105 3e-23
tgo:TGME49_035540 translation initiation factor 2 beta, putati... 94.7 6e-20
cpv:cgd8_4430 translation initiation factor if-2 betam beta su... 92.8 3e-19
bbo:BBOV_II007040 18.m06583; eukaryotic translation initiation... 89.7 2e-18
pfa:PF10_0103 eukaryotic translation initiation factor 2 beta,... 84.3 9e-17
mmu:100040220 Gm9892; predicted gene 9892 54.3
mmu:67204 Eif2s2, 2810026E11Rik, 38kDa, AA408636, AA571381, AA... 54.3 1e-07
hsa:8894 EIF2S2, DKFZp686L18198, EIF2, EIF2B, EIF2beta, MGC850... 53.9 1e-07
xla:779195 hypothetical protein MGC130708 52.8 3e-07
dre:324365 eif2s2, wu:fc26g03, wu:fu10f07, zgc:77084; eukaryot... 52.8 3e-07
xla:779197 eif2s2, MGC130959; eukaryotic translation initiatio... 52.8 3e-07
xla:780741 eukaryote initiation factor 2 beta 52.8 3e-07
ath:AT5G20920 EIF2 BETA; translation initiation factor; K03238... 50.4 1e-06
cel:K04G2.1 iftb-1; eIFTwoBeta (eIF2beta translation initiatio... 44.3 8e-05
sce:YPL237W SUI3; Beta subunit of the translation initiation f... 36.2 0.023
cel:Y54E10A.4 fog-1; Feminization Of Germline family member (f... 32.3 0.34
dre:555994 gramd1a, si:ch211-225p5.11, wu:fc88h04; GRAM domain... 30.4 1.4
ath:AT5G22810 GDSL-motif lipase, putative 29.3 3.4
mmu:640524 Spnb5, EG640524, Gm354; spectrin beta 5; K06115 spe... 28.1 7.4
cel:F15A8.6 hypothetical protein; K01044 carboxylesterase [EC:... 27.7 9.4
> tpv:TP02_0678 eukaryotic translation initiation factor 2 subunit
beta; K03238 translation initiation factor 2 subunit 2
Length=241
Score = 105 bits (262), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 52/81 (64%), Positives = 65/81 (80%), Gaps = 5/81 (6%)
Query 27 FDFGERRKKKKEKKQEDQGEAAAAEGFIDGSGQIFVRGNVYTYEEMLSRIQDLIVKNNPD 86
F+FGE KK+K+ D G++ E IDG+GQ+FV+G+VY YEE+L+RIQ +I NNPD
Sbjct 49 FNFGE----KKKKRTVDAGDSPKTE-IIDGTGQVFVKGHVYPYEELLARIQSMINANNPD 103
Query 87 LAGSKRYTIKPPQVVRVGSKK 107
L+GSKRYTIKPPQVVRVGSKK
Sbjct 104 LSGSKRYTIKPPQVVRVGSKK 124
> tgo:TGME49_035540 translation initiation factor 2 beta, putative
; K03238 translation initiation factor 2 subunit 2
Length=231
Score = 94.7 bits (234), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 59/96 (61%), Positives = 75/96 (78%), Gaps = 6/96 (6%)
Query 17 AEAQQP-----AAQAFDFGERRKKKKEKKQEDQGEAAAAEGFIDGSGQIFVRGNVYTYEE 71
AEA+QP A Q FDFGERR++KK++K+E + +A IDG+G++F RG Y+Y+E
Sbjct 19 AEAKQPEGEEEAPQQFDFGERRRRKKKEKKETE-QAQEEAPIIDGTGELFKRGKEYSYQE 77
Query 72 MLSRIQDLIVKNNPDLAGSKRYTIKPPQVVRVGSKK 107
ML RIQ LI K+NPDL+G+KRYTIKPPQVVRVGSKK
Sbjct 78 MLQRIQTLIEKHNPDLSGAKRYTIKPPQVVRVGSKK 113
> cpv:cgd8_4430 translation initiation factor if-2 betam beta
subunit ZnR ; K03238 translation initiation factor 2 subunit
2
Length=214
Score = 92.8 bits (229), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 49/90 (54%), Positives = 67/90 (74%), Gaps = 1/90 (1%)
Query 18 EAQQPAAQAFDFGERRKKKKEKKQEDQGEAAAAEGFIDGSGQIFVRGNVYTYEEMLSRIQ 77
E Q +DFGE+ KKKK+ K+ G + ++DGSGQ+FV+G +Y YEE+L R++
Sbjct 8 EGVQKMDMGYDFGEK-KKKKKSKEHKTGVSEVESSYVDGSGQLFVKGAIYPYEELLERVR 66
Query 78 DLIVKNNPDLAGSKRYTIKPPQVVRVGSKK 107
LI+++NPDL G+KRYT+KPPQVVRVGSKK
Sbjct 67 RLILEHNPDLWGAKRYTLKPPQVVRVGSKK 96
> bbo:BBOV_II007040 18.m06583; eukaryotic translation initiation
factor 2, beta; K03238 translation initiation factor 2 subunit
2
Length=238
Score = 89.7 bits (221), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 52/98 (53%), Positives = 67/98 (68%), Gaps = 9/98 (9%)
Query 10 APDTAATAEAQQPAAQAFDFGERRKKKKEKKQEDQGEAAAAEGFIDGSGQIFVRGNVYTY 69
AP A AE A +DFGE++KKK + + IDGSGQ+FVRG+VY Y
Sbjct 33 APAAAVKAEG----APKYDFGEKKKKKAAPASD-----SVKSEVIDGSGQVFVRGHVYQY 83
Query 70 EEMLSRIQDLIVKNNPDLAGSKRYTIKPPQVVRVGSKK 107
EE+L+RIQ LI ++ DL+G+K+YTI+PPQVVRVGSKK
Sbjct 84 EELLNRIQTLINAHSHDLSGNKKYTIRPPQVVRVGSKK 121
> pfa:PF10_0103 eukaryotic translation initiation factor 2 beta,
putative; K03238 translation initiation factor 2 subunit
2
Length=222
Score = 84.3 bits (207), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 51/85 (60%), Positives = 65/85 (76%), Gaps = 4/85 (4%)
Query 23 AAQAFDFGERRKKKKEKKQEDQGEAAAAEGFIDGSGQIFVRGNVYTYEEMLSRIQDLIVK 82
+ Q FDFGE++KKKK+K+ ++ E IDG+G++F RG VY Y+E+L RIQDLI K
Sbjct 23 SKQLFDFGEKKKKKKKKEVVEKVEEI----IIDGTGKVFERGAVYPYDELLHRIQDLINK 78
Query 83 NNPDLAGSKRYTIKPPQVVRVGSKK 107
+N DL SK+YTIKPPQVVRVGSKK
Sbjct 79 HNIDLCISKKYTIKPPQVVRVGSKK 103
> mmu:100040220 Gm9892; predicted gene 9892
Length=331
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 24/43 (55%), Positives = 36/43 (83%), Gaps = 2/43 (4%)
Query 67 YTYEEMLSRIQDLIVKNNPDL-AGSKR-YTIKPPQVVRVGSKK 107
YTYEE+L+R+ +++ + NPD+ AG KR + +KPPQV+RVG+KK
Sbjct 172 YTYEELLNRVLNIMREKNPDMVAGEKRKFVMKPPQVIRVGTKK 214
> mmu:67204 Eif2s2, 2810026E11Rik, 38kDa, AA408636, AA571381,
AA986487, AW822225, D2Ertd303e, EIF2, EIF2B, MGC130606; eukaryotic
translation initiation factor 2, subunit 2 (beta); K03238
translation initiation factor 2 subunit 2
Length=331
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 25/43 (58%), Positives = 36/43 (83%), Gaps = 2/43 (4%)
Query 67 YTYEEMLSRIQDLIVKNNPDL-AGSKR-YTIKPPQVVRVGSKK 107
YTYEE+L+R+ +++ + NPD+ AG KR + +KPPQVVRVG+KK
Sbjct 172 YTYEELLNRVFNIMREKNPDMVAGEKRKFVMKPPQVVRVGTKK 214
> hsa:8894 EIF2S2, DKFZp686L18198, EIF2, EIF2B, EIF2beta, MGC8508;
eukaryotic translation initiation factor 2, subunit 2 beta,
38kDa; K03238 translation initiation factor 2 subunit 2
Length=333
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 25/43 (58%), Positives = 36/43 (83%), Gaps = 2/43 (4%)
Query 67 YTYEEMLSRIQDLIVKNNPDL-AGSKR-YTIKPPQVVRVGSKK 107
YTYEE+L+R+ +++ + NPD+ AG KR + +KPPQVVRVG+KK
Sbjct 174 YTYEELLNRVFNIMREKNPDMVAGEKRKFVMKPPQVVRVGTKK 216
> xla:779195 hypothetical protein MGC130708
Length=332
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 24/43 (55%), Positives = 36/43 (83%), Gaps = 2/43 (4%)
Query 67 YTYEEMLSRIQDLIVKNNPDL-AGSKR-YTIKPPQVVRVGSKK 107
YTY+E+L+R+ +++ + NPD+ AG KR + +KPPQVVRVG+KK
Sbjct 173 YTYDELLNRVFNIMREKNPDMVAGEKRKFVMKPPQVVRVGTKK 215
> dre:324365 eif2s2, wu:fc26g03, wu:fu10f07, zgc:77084; eukaryotic
translation initiation factor 2, subunit 2 beta; K03238
translation initiation factor 2 subunit 2
Length=327
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 24/43 (55%), Positives = 36/43 (83%), Gaps = 2/43 (4%)
Query 67 YTYEEMLSRIQDLIVKNNPDL-AGSKR-YTIKPPQVVRVGSKK 107
YTY+E+L+R+ +++ + NPD+ AG KR + +KPPQVVRVG+KK
Sbjct 168 YTYDELLNRVFNIMREKNPDMVAGEKRKFVMKPPQVVRVGTKK 210
> xla:779197 eif2s2, MGC130959; eukaryotic translation initiation
factor 2, subunit 2 beta, 38kDa; K03238 translation initiation
factor 2 subunit 2
Length=335
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 24/43 (55%), Positives = 36/43 (83%), Gaps = 2/43 (4%)
Query 67 YTYEEMLSRIQDLIVKNNPDL-AGSKR-YTIKPPQVVRVGSKK 107
YTY+E+L+R+ +++ + NPD+ AG KR + +KPPQVVRVG+KK
Sbjct 176 YTYDELLNRVFNIMREKNPDMVAGEKRKFVMKPPQVVRVGTKK 218
> xla:780741 eukaryote initiation factor 2 beta
Length=335
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 24/43 (55%), Positives = 36/43 (83%), Gaps = 2/43 (4%)
Query 67 YTYEEMLSRIQDLIVKNNPDL-AGSKR-YTIKPPQVVRVGSKK 107
YTY+E+L+R+ +++ + NPD+ AG KR + +KPPQVVRVG+KK
Sbjct 176 YTYDELLNRVFNIMREKNPDMVAGEKRKFVMKPPQVVRVGTKK 218
> ath:AT5G20920 EIF2 BETA; translation initiation factor; K03238
translation initiation factor 2 subunit 2
Length=268
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 21/42 (50%), Positives = 34/42 (80%), Gaps = 1/42 (2%)
Query 67 YTYEEMLSRIQDLIVKNNPDLAGSKRYTI-KPPQVVRVGSKK 107
Y Y+E+L R+ +++ +NNP+LAG +R T+ +PPQV+R G+KK
Sbjct 116 YIYDELLGRVFNILRENNPELAGDRRRTVMRPPQVLREGTKK 157
> cel:K04G2.1 iftb-1; eIFTwoBeta (eIF2beta translation initiation
factor) family member (iftb-1); K03238 translation initiation
factor 2 subunit 2
Length=250
Score = 44.3 bits (103), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 21/42 (50%), Positives = 28/42 (66%), Gaps = 1/42 (2%)
Query 67 YTYEEMLSRIQDLIVKNNPDLAGS-KRYTIKPPQVVRVGSKK 107
YTYEE L+ + ++ NPD AG K++ IK P+V R GSKK
Sbjct 87 YTYEEALTLVYQVMKDKNPDFAGDKKKFAIKLPEVARAGSKK 128
> sce:YPL237W SUI3; Beta subunit of the translation initiation
factor eIF2, involved in the identification of the start codon;
proposed to be involved in mRNA binding; K03238 translation
initiation factor 2 subunit 2
Length=285
Score = 36.2 bits (82), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 17/41 (41%), Positives = 26/41 (63%), Gaps = 3/41 (7%)
Query 69 YEEMLSRIQDLIVKNNPDLAGSK---RYTIKPPQVVRVGSK 106
Y E+LSR +++ NNP+LAG + ++ I PP +R G K
Sbjct 131 YSELLSRFFNILRTNNPELAGDRSGPKFRIPPPVCLRDGKK 171
> cel:Y54E10A.4 fog-1; Feminization Of Germline family member
(fog-1)
Length=619
Score = 32.3 bits (72), Expect = 0.34, Method: Composition-based stats.
Identities = 23/94 (24%), Positives = 41/94 (43%), Gaps = 9/94 (9%)
Query 3 CCSPPPPAPDTAATAEAQQPAAQAFDFGERRKKKKEKKQ---EDQGEAAAAEGFIDGSGQ 59
CC+P PP + Q PAA DFG + + E + G+
Sbjct 121 CCNPAPPY-----ASHCQSPAASD-DFGHHFGSFDSMSAYNFHNNNQQQRREAHVSRDGK 174
Query 60 IFVRGNVYTYEEMLSRIQDLIVKNNPDLAGSKRY 93
F +GN+YT E++ + +++ +NN + +K +
Sbjct 175 SFDQGNIYTAEDVHTVVKNHRNENNQRVVSNKVF 208
> dre:555994 gramd1a, si:ch211-225p5.11, wu:fc88h04; GRAM domain
containing 1A
Length=789
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 13/40 (32%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 8 PPAPDTAATAEAQQPAAQAFDFGERRKKKKEKKQEDQGEA 47
PP+ DT + + + P A + R K+K + +QED+ A
Sbjct 24 PPSSDTESWSRSPSPRAHRYRSRSREKRKAKAQQEDKSGA 63
> ath:AT5G22810 GDSL-motif lipase, putative
Length=337
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 13/42 (30%), Positives = 26/42 (61%), Gaps = 4/42 (9%)
Query 47 AAAAEGFIDGSGQIF----VRGNVYTYEEMLSRIQDLIVKNN 84
A+AA G+ DG+ +++ + + Y++ +SRIQ++ NN
Sbjct 93 ASAASGYYDGTAKLYSAISLPQQLEHYKDYISRIQEIATSNN 134
> mmu:640524 Spnb5, EG640524, Gm354; spectrin beta 5; K06115 spectrin
beta
Length=3663
Score = 28.1 bits (61), Expect = 7.4, Method: Composition-based stats.
Identities = 20/81 (24%), Positives = 40/81 (49%), Gaps = 6/81 (7%)
Query 10 APDTAATAEAQQPAAQAFDFGE-----RRKKKKEKKQEDQGEA-AAAEGFIDGSGQIFVR 63
A + A + + P+A A D+G+ R ++K + E++ + A + G+G ++
Sbjct 2926 ADEEMAWVQEKLPSATAQDYGQSLNTVRHLQEKHQNLENEIHSHKALSQVVTGTGHKLIQ 2985
Query 64 GNVYTYEEMLSRIQDLIVKNN 84
+ EE+ +R+Q L V N
Sbjct 2986 AGHFATEEVAARVQQLEVALN 3006
> cel:F15A8.6 hypothetical protein; K01044 carboxylesterase [EC:3.1.1.1]
Length=565
Score = 27.7 bits (60), Expect = 9.4, Method: Composition-based stats.
Identities = 16/60 (26%), Positives = 27/60 (45%), Gaps = 1/60 (1%)
Query 36 KKEKKQEDQGEAAAAEGFIDGSGQIFVRGNVYTYEEMLSRIQDLIVKNNPDLAGSKRYTI 95
KK ED E AA G+I +FV Y Y E ++++ + + + D + + I
Sbjct 396 KKLSDDEDDKEVAA-RGYIQLYSDLFVNNGTYNYAEKMTKLGNKVFMYSFDYCNPRSFGI 454
Lambda K H
0.312 0.130 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2007980300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40