bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
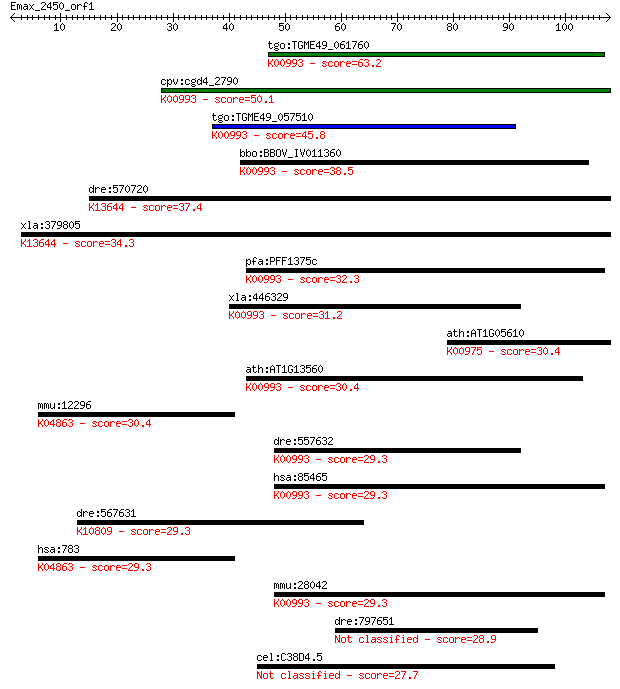
Query= Emax_2450_orf1
Length=107
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_061760 ethanolaminephosphotransferase, putative (EC... 63.2 2e-10
cpv:cgd4_2790 ethanolaminephosphotransferase (ETHPT) 9 transme... 50.1 2e-06
tgo:TGME49_057510 aminoalcoholphosphotransferase, putative (EC... 45.8 3e-05
bbo:BBOV_IV011360 23.m05792; ethanolamine phosphatidyltransfer... 38.5 0.005
dre:570720 cept1, zgc:171762; choline/ethanolamine phosphotran... 37.4 0.012
xla:379805 cept1, MGC53412; choline/ethanolamine phosphotransf... 34.3 0.092
pfa:PFF1375c ethanolaminephosphotransferase, putative (EC:2.7.... 32.3 0.37
xla:446329 ept1, MGC78842, seli, sepi; ethanolaminephosphotran... 31.2 0.86
ath:AT1G05610 APS2; APS2 (ADP-glucose pyrophoshorylase small s... 30.4 1.3
ath:AT1G13560 AAPT1; AAPT1 (AMINOALCOHOLPHOSPHOTRANSFERASE 1);... 30.4 1.3
mmu:12296 Cacnb2, AW060387, Cavbeta2, Cchb2, MGC129334, MGC129... 30.4 1.5
dre:557632 choline/ethanolaminephosphotransferase-like; K00993... 29.3 2.8
hsa:85465 EPT1, KIAA1724, SELI, SEPI; ethanolaminephosphotrans... 29.3 2.8
dre:567631 tg, cb717; thyroglobulin; K10809 thyroglobulin 29.3 3.1
hsa:783 CACNB2, CACNLB2, CAVB2, FLJ23743, MYSB; calcium channe... 29.3 3.1
mmu:28042 Ept1, 4933402G07Rik, AI448296, AI452230, C79563, D5W... 29.3 3.5
dre:797651 jam3a, jamc.2, si:ch211-277g22.3; junctional adhesi... 28.9 4.3
cel:C38D4.5 tag-325; Temporarily Assigned Gene name family mem... 27.7 9.1
> tgo:TGME49_061760 ethanolaminephosphotransferase, putative (EC:2.7.8.1);
K00993 ethanolaminephosphotransferase [EC:2.7.8.1]
Length=449
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 30/60 (50%), Positives = 36/60 (60%), Gaps = 0/60 (0%)
Query 47 GVASLHKYAYVSGHRTPFDTIMNPWWEAVEKLVPRCVHPSALTVSSCTAAIIGSCLLLGF 106
G+ +LH Y Y SG TP D +MNPWWE V LVP VHP+ LTV AI + L L +
Sbjct 11 GLKNLHSYKYSSGGYTPLDKVMNPWWEFVASLVPPTVHPNVLTVVGFLCAIGAAVLQLTY 70
> cpv:cgd4_2790 ethanolaminephosphotransferase (ETHPT) 9 transmembrane
domain protein involved in lipid ; K00993 ethanolaminephosphotransferase
[EC:2.7.8.1]
Length=428
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 23/80 (28%), Positives = 42/80 (52%), Gaps = 1/80 (1%)
Query 28 TRRRLQHNEGGFWALLDAAGVASLHKYAYVSGHRTPFDTIMNPWWEAVEKLVPRCVHPSA 87
++ ++ H G F + + G+ ++ +Y+Y SG T D MNP+WE +P C+ P+
Sbjct 5 SKEKITH-VGVFGSFISEVGLKNIKEYSYKSGGVTFLDYAMNPFWEFFAHQIPECISPNL 63
Query 88 LTVSSCTAAIIGSCLLLGFC 107
LT+ ++I L + C
Sbjct 64 LTIFGFLCSLIAMLLTMMTC 83
> tgo:TGME49_057510 aminoalcoholphosphotransferase, putative (EC:2.7.8.1);
K00993 ethanolaminephosphotransferase [EC:2.7.8.1]
Length=467
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 21/54 (38%), Positives = 29/54 (53%), Gaps = 0/54 (0%)
Query 37 GGFWALLDAAGVASLHKYAYVSGHRTPFDTIMNPWWEAVEKLVPRCVHPSALTV 90
G F + G+ L +Y Y G TP D +MNPWW+ V +P + P+ LTV
Sbjct 5 GVFGCFITRQGLEELRRYEYRGGGSTPIDRLMNPWWDYVAGKLPLWLSPNLLTV 58
> bbo:BBOV_IV011360 23.m05792; ethanolamine phosphatidyltransferase
(EC:2.7.8.1); K00993 ethanolaminephosphotransferase [EC:2.7.8.1]
Length=401
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 33/64 (51%), Gaps = 2/64 (3%)
Query 42 LLDAAGVASLHKYAYVSGHRTPFDTIMNP-WWEAVEKLVPRCVHPSALTVSSCTAAI-IG 99
++ A + L Y + SG T D ++N WW + L+PR + P+ +T+S I +
Sbjct 7 MIPKANMPKLKDYKFQSGGSTKLDCLINVIWWNPIASLIPRFISPNVITLSGTLCLIAMN 66
Query 100 SCLL 103
C+L
Sbjct 67 YCIL 70
> dre:570720 cept1, zgc:171762; choline/ethanolamine phosphotransferase
1; K13644 choline/ethanolamine phosphotransferase
[EC:2.7.8.1 2.7.8.2]
Length=415
Score = 37.4 bits (85), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 23/93 (24%), Positives = 43/93 (46%), Gaps = 16/93 (17%)
Query 15 LLRRYSELGGPLLTRRRLQHNEGGFWALLDAAGVASLHKYAYVSGHRTPFDTIMNPWWEA 74
+LRR EL P+L+R +L+ L ++ Y S R+ + IM +WE
Sbjct 31 VLRRLFELPAPVLSRHQLKR----------------LEEHRYSSSGRSLLEPIMQQYWEW 74
Query 75 VEKLVPRCVHPSALTVSSCTAAIIGSCLLLGFC 107
+ +P + P+ +T+ I + +L+ +C
Sbjct 75 LVNRMPPWIAPNLITIVGLVTNIFTTLVLVYYC 107
> xla:379805 cept1, MGC53412; choline/ethanolamine phosphotransferase
1 (EC:2.7.8.2 2.7.8.1); K13644 choline/ethanolamine
phosphotransferase [EC:2.7.8.1 2.7.8.2]
Length=416
Score = 34.3 bits (77), Expect = 0.092, Method: Composition-based stats.
Identities = 23/105 (21%), Positives = 47/105 (44%), Gaps = 16/105 (15%)
Query 3 PGICMHRRGRHTLLRRYSELGGPLLTRRRLQHNEGGFWALLDAAGVASLHKYAYVSGHRT 62
P C + LL ++ EL P L+R +L+ L ++ Y S ++
Sbjct 19 PSSCGNPYQTACLLSKFIELPNPPLSRHQLKR----------------LEEHRYQSCGKS 62
Query 63 PFDTIMNPWWEAVEKLVPRCVHPSALTVSSCTAAIIGSCLLLGFC 107
+ +M +WE + VP+ + P+ +T+ I+ + +L+ +C
Sbjct 63 LLEPLMQGFWEWLVIQVPQWIAPNLITIIGLLINIVTTVVLIYYC 107
> pfa:PFF1375c ethanolaminephosphotransferase, putative (EC:2.7.8.1);
K00993 ethanolaminephosphotransferase [EC:2.7.8.1]
Length=391
Score = 32.3 bits (72), Expect = 0.37, Method: Composition-based stats.
Identities = 16/64 (25%), Positives = 29/64 (45%), Gaps = 0/64 (0%)
Query 43 LDAAGVASLHKYAYVSGHRTPFDTIMNPWWEAVEKLVPRCVHPSALTVSSCTAAIIGSCL 102
L+ + ++ Y Y SG + D + + +W K VP+ + P+ LT+ + I L
Sbjct 7 LNKSVYSNCKSYVYKSGGNSLLDNLFDAFWNICVKFVPKSITPNLLTLLGFLCSTIAFVL 66
Query 103 LLGF 106
F
Sbjct 67 TFVF 70
> xla:446329 ept1, MGC78842, seli, sepi; ethanolaminephosphotransferase
1 (CDP-ethanolamine-specific) (EC:2.7.8.1); K00993
ethanolaminephosphotransferase [EC:2.7.8.1]
Length=401
Score = 31.2 bits (69), Expect = 0.86, Method: Composition-based stats.
Identities = 13/53 (24%), Positives = 25/53 (47%), Gaps = 1/53 (1%)
Query 40 WALLDAAGVASLHKYAYVSGHRTPFDT-IMNPWWEAVEKLVPRCVHPSALTVS 91
W + +A KY Y + P +M+P+W ++ + P + P+ +T S
Sbjct 3 WEYVSPEQLAGFDKYKYSAVDTNPLSLYVMHPFWNSIVRFFPTWLAPNLITFS 55
> ath:AT1G05610 APS2; APS2 (ADP-glucose pyrophoshorylase small
subunit 2); glucose-1-phosphate adenylyltransferase/ nucleotidyltransferase/
transferase (EC:2.7.7.27); K00975 glucose-1-phosphate
adenylyltransferase [EC:2.7.7.27]
Length=476
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 12/29 (41%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 79 VPRCVHPSALTVSSCTAAIIGSCLLLGFC 107
+PRC+ PS+++V+ T +IIG +L C
Sbjct 337 MPRCLPPSSMSVAVITNSIIGDGCILDKC 365
> ath:AT1G13560 AAPT1; AAPT1 (AMINOALCOHOLPHOSPHOTRANSFERASE 1);
phosphatidyltransferase/ phosphotransferase, for other substituted
phosphate groups (EC:2.7.8.1); K00993 ethanolaminephosphotransferase
[EC:2.7.8.1]
Length=389
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 31/62 (50%), Gaps = 3/62 (4%)
Query 43 LDAAGVASLHKYAYVSG--HRTPFDTIMNPWWEAVEKLVPRCVHPSALTVSSCTAAIIGS 100
+ A GVA+LH+Y Y SG H ++ P+W K+ P + P+ +T+ + S
Sbjct 4 IGAHGVAALHRYKY-SGVDHSYLAKYVLQPFWTRFVKVFPLWMPPNMITLMGFMFLVTSS 62
Query 101 CL 102
L
Sbjct 63 LL 64
> mmu:12296 Cacnb2, AW060387, Cavbeta2, Cchb2, MGC129334, MGC129335;
calcium channel, voltage-dependent, beta 2 subunit; K04863
voltage-dependent calcium channel beta-2
Length=655
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 20/35 (57%), Gaps = 3/35 (8%)
Query 6 CMHRRGRHTLLRRYSELGGPLLTRRRLQHNEGGFW 40
C+ +R RH RY + G ++++RR NE G W
Sbjct 616 CIKQRSRHKSKDRYCDKEGEVISKRR---NEAGEW 647
> dre:557632 choline/ethanolaminephosphotransferase-like; K00993
ethanolaminephosphotransferase [EC:2.7.8.1]
Length=386
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 12/45 (26%), Positives = 25/45 (55%), Gaps = 1/45 (2%)
Query 48 VASLHKYAYVSGHRTPFDT-IMNPWWEAVEKLVPRCVHPSALTVS 91
++ KY Y + P +M+P+W +V K++P + P+ +T +
Sbjct 12 LSGFDKYKYSAVDSNPLSIYVMHPFWNSVVKVLPTWLAPNLITFT 56
> hsa:85465 EPT1, KIAA1724, SELI, SEPI; ethanolaminephosphotransferase
1 (CDP-ethanolamine-specific) (EC:2.7.8.1); K00993
ethanolaminephosphotransferase [EC:2.7.8.1]
Length=397
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 15/60 (25%), Positives = 27/60 (45%), Gaps = 1/60 (1%)
Query 48 VASLHKYAYVSGHRTPFDT-IMNPWWEAVEKLVPRCVHPSALTVSSCTAAIIGSCLLLGF 106
+A KY Y + P +M+P+W + K+ P + P+ +T S + L+ F
Sbjct 12 LAGFDKYKYSAVDTNPLSLYVMHPFWNTIVKVFPTWLAPNLITFSGFLLVVFNFLLMAYF 71
> dre:567631 tg, cb717; thyroglobulin; K10809 thyroglobulin
Length=2539
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 18/56 (32%), Positives = 27/56 (48%), Gaps = 5/56 (8%)
Query 13 HTLLRRYSELGGPLLTRRRLQHNEGGFWALLDAAGVASLHKY-----AYVSGHRTP 63
H R Y EL L+ R+ LQ ++ FW+ A +S K+ A +G +TP
Sbjct 2457 HVGGRAYKELSSTLVNRKNLQRSQCSFWSQYVPALTSSTAKFSCGTSAEETGVQTP 2512
> hsa:783 CACNB2, CACNLB2, CAVB2, FLJ23743, MYSB; calcium channel,
voltage-dependent, beta 2 subunit; K04863 voltage-dependent
calcium channel beta-2
Length=605
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 19/35 (54%), Gaps = 3/35 (8%)
Query 6 CMHRRGRHTLLRRYSELGGPLLTRRRLQHNEGGFW 40
C +R RH RY E G +++++R NE G W
Sbjct 566 CNKQRSRHKSKDRYCEKDGEVISKKR---NEAGEW 597
> mmu:28042 Ept1, 4933402G07Rik, AI448296, AI452230, C79563, D5Wsu178e,
MGC117838, SELI, mKIAA1724; ethanolaminephosphotransferase
1 (CDP-ethanolamine-specific) (EC:2.7.8.1); K00993
ethanolaminephosphotransferase [EC:2.7.8.1]
Length=398
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 16/60 (26%), Positives = 27/60 (45%), Gaps = 1/60 (1%)
Query 48 VASLHKYAYVSGHRTPFDT-IMNPWWEAVEKLVPRCVHPSALTVSSCTAAIIGSCLLLGF 106
++ KY Y + P IM+P+W + K+ P + P+ +T S + LL F
Sbjct 12 LSGFDKYKYSALDTNPLSLYIMHPFWNTIVKVFPTWLAPNLITFSGFMLLVFNFLLLTYF 71
> dre:797651 jam3a, jamc.2, si:ch211-277g22.3; junctional adhesion
molecule 3a
Length=301
Score = 28.9 bits (63), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 19/36 (52%), Gaps = 0/36 (0%)
Query 59 GHRTPFDTIMNPWWEAVEKLVPRCVHPSALTVSSCT 94
+ PFD I+ V+ ++PRC P A+ V S T
Sbjct 113 DDQKPFDEILISLAVRVKPVIPRCSVPDAVNVGSST 148
> cel:C38D4.5 tag-325; Temporarily Assigned Gene name family member
(tag-325)
Length=837
Score = 27.7 bits (60), Expect = 9.1, Method: Composition-based stats.
Identities = 18/53 (33%), Positives = 24/53 (45%), Gaps = 3/53 (5%)
Query 45 AAGVASLHKYAYVSGHRTPFDTIMNPWWEAVEKLVPRCVHPSALTVSSCTAAI 97
A+GV H YA+VS ++ F N E +KL + P A S AI
Sbjct 256 ASGVMMPHPYAHVSSFQSTFPRAKN---EDTDKLASYVIPPDANETSKTPTAI 305
Lambda K H
0.327 0.140 0.477
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2007980300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40