bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2419_orf1
Length=112
Score E
Sequences producing significant alignments: (Bits) Value
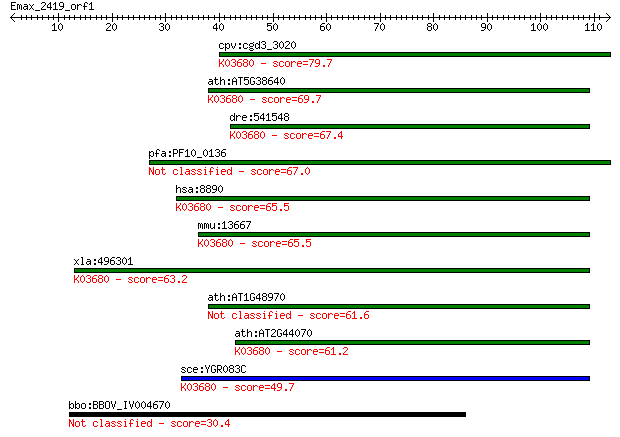
cpv:cgd3_3020 possible guaning nucleotide exchange factor, eIF... 79.7 2e-15
ath:AT5G38640 eukaryotic translation initiation factor 2B fami... 69.7 2e-12
dre:541548 eif2b4, si:ch211-101l18.4, wu:fb54a09, zgc:110790; ... 67.4 1e-11
pfa:PF10_0136 initiation factor 2 subunit family, putative 67.0 2e-11
hsa:8890 EIF2B4, DKFZp586J0119, EIF-2B, EIF2B, EIF2Bdelta; euk... 65.5 4e-11
mmu:13667 Eif2b4, Eif2b; eukaryotic translation initiation fac... 65.5 5e-11
xla:496301 eif2b4; eukaryotic translation initiation factor 2B... 63.2 2e-10
ath:AT1G48970 GTP binding / translation initiation factor 61.6 6e-10
ath:AT2G44070 GTP binding / translation initiation factor; K03... 61.2 8e-10
sce:YGR083C GCD2, GCD12; Delta subunit of the translation init... 49.7 2e-06
bbo:BBOV_IV004670 23.m06553; hypothetical protein 30.4 1.3
> cpv:cgd3_3020 possible guaning nucleotide exchange factor, eIF-2B
; K03680 translation initiation factor eIF-2B subunit
delta
Length=357
Score = 79.7 bits (195), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 34/73 (46%), Positives = 52/73 (71%), Gaps = 0/73 (0%)
Query 40 QEIIHPAVIRVGLQMANRKIQGANARTVAMLSSFNCFIEDYSPPPYESIDRHIKLSLDRR 99
Q IHPA++R+G++M +RKI G N R+ +L + +EDY PPY+SI++H+K+ LD
Sbjct 34 QSKIHPAILRLGMRMKDRKIVGTNMRSKCLLIAIGQMLEDYYCPPYKSIEKHLKMVLDVH 93
Query 100 INFITNCRPHNFA 112
I++IT+ R HN A
Sbjct 94 ISYITSQRQHNIA 106
> ath:AT5G38640 eukaryotic translation initiation factor 2B family
protein / eIF-2B family protein; K03680 translation initiation
factor eIF-2B subunit delta
Length=642
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 29/71 (40%), Positives = 46/71 (64%), Gaps = 0/71 (0%)
Query 38 FRQEIIHPAVIRVGLQMANRKIQGANARTVAMLSSFNCFIEDYSPPPYESIDRHIKLSLD 97
F+ + +HPAV +VGLQ + I G NAR +AML +F ++DYS PP +S++R + +
Sbjct 312 FQLDPMHPAVYKVGLQYLSGDISGGNARCIAMLQAFQEVVKDYSTPPEKSLNRDMTAKIS 371
Query 98 RRINFITNCRP 108
++F+ CRP
Sbjct 372 SYVSFLIECRP 382
> dre:541548 eif2b4, si:ch211-101l18.4, wu:fb54a09, zgc:110790;
eukaryotic translation initiation factor 2B, subunit 4 delta;
K03680 translation initiation factor eIF-2B subunit delta
Length=540
Score = 67.4 bits (163), Expect = 1e-11, Method: Composition-based stats.
Identities = 30/67 (44%), Positives = 46/67 (68%), Gaps = 0/67 (0%)
Query 42 IIHPAVIRVGLQMANRKIQGANARTVAMLSSFNCFIEDYSPPPYESIDRHIKLSLDRRIN 101
+IHP+++R+GLQ + I G+NAR+VA+L +F I+DYS PP E + R + L I+
Sbjct 219 VIHPSIVRLGLQYSQGIIAGSNARSVALLHAFKQVIQDYSTPPNEELSRDLVNKLSPYIS 278
Query 102 FITNCRP 108
F++ CRP
Sbjct 279 FLSQCRP 285
> pfa:PF10_0136 initiation factor 2 subunit family, putative
Length=1074
Score = 67.0 bits (162), Expect = 2e-11, Method: Composition-based stats.
Identities = 29/86 (33%), Positives = 54/86 (62%), Gaps = 0/86 (0%)
Query 27 RLPLHLRLLMNFRQEIIHPAVIRVGLQMANRKIQGANARTVAMLSSFNCFIEDYSPPPYE 86
++ ++ ++L++ Q IHP ++R G+ I N R V +L + FI+DY+ PPYE
Sbjct 307 KINMYDKILLDLNQNEIHPNILRTGIFFNKCSITTHNHRNVDLLIALKSFIKDYTLPPYE 366
Query 87 SIDRHIKLSLDRRINFITNCRPHNFA 112
I++H+K+ +D+ IN+I C+ H+ +
Sbjct 367 PINKHMKVVIDKEINYIIMCKKHSVS 392
> hsa:8890 EIF2B4, DKFZp586J0119, EIF-2B, EIF2B, EIF2Bdelta; eukaryotic
translation initiation factor 2B, subunit 4 delta,
67kDa; K03680 translation initiation factor eIF-2B subunit
delta
Length=523
Score = 65.5 bits (158), Expect = 4e-11, Method: Composition-based stats.
Identities = 29/77 (37%), Positives = 47/77 (61%), Gaps = 0/77 (0%)
Query 32 LRLLMNFRQEIIHPAVIRVGLQMANRKIQGANARTVAMLSSFNCFIEDYSPPPYESIDRH 91
L M+ +IHPA++R+GLQ + + G+NAR +A+L + I+DY+ PP E + R
Sbjct 192 LTQFMSIPSSVIHPAMVRLGLQYSQGLVSGSNARCIALLRALQQVIQDYTTPPNEELSRD 251
Query 92 IKLSLDRRINFITNCRP 108
+ L ++F+T CRP
Sbjct 252 LVNKLKPYMSFLTQCRP 268
> mmu:13667 Eif2b4, Eif2b; eukaryotic translation initiation factor
2B, subunit 4 delta; K03680 translation initiation factor
eIF-2B subunit delta
Length=544
Score = 65.5 bits (158), Expect = 5e-11, Method: Composition-based stats.
Identities = 30/73 (41%), Positives = 46/73 (63%), Gaps = 0/73 (0%)
Query 36 MNFRQEIIHPAVIRVGLQMANRKIQGANARTVAMLSSFNCFIEDYSPPPYESIDRHIKLS 95
M+ +IHPA++R+GLQ + I G+NAR +A+L + I+DY+ PP E + R +
Sbjct 217 MSIPSSVIHPAMVRLGLQYSQGLISGSNARCIALLHALQQVIQDYTTPPSEELSRDLVNK 276
Query 96 LDRRINFITNCRP 108
L I+F+T CRP
Sbjct 277 LKPYISFLTQCRP 289
> xla:496301 eif2b4; eukaryotic translation initiation factor
2B, subunit 4 delta, 67kDa; K03680 translation initiation factor
eIF-2B subunit delta
Length=569
Score = 63.2 bits (152), Expect = 2e-10, Method: Composition-based stats.
Identities = 35/96 (36%), Positives = 53/96 (55%), Gaps = 2/96 (2%)
Query 13 TIISSSSSSNSSSSRLPLHLRLLMNFRQEIIHPAVIRVGLQMANRKIQGANARTVAMLSS 72
T +S S + S +LPL M+ +IHP V+R+GLQ ++ I G+N+R +A+L
Sbjct 221 TKVSLFSHLHQYSRKLPLTQH--MSIPSSVIHPVVVRLGLQYSHGIINGSNSRCIALLHC 278
Query 73 FNCFIEDYSPPPYESIDRHIKLSLDRRINFITNCRP 108
F I DY+ P E + R + L I+F+ CRP
Sbjct 279 FKQVIRDYTTPQQEELSRDLVNKLKPYISFLNQCRP 314
> ath:AT1G48970 GTP binding / translation initiation factor
Length=666
Score = 61.6 bits (148), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 29/71 (40%), Positives = 41/71 (57%), Gaps = 0/71 (0%)
Query 38 FRQEIIHPAVIRVGLQMANRKIQGANARTVAMLSSFNCFIEDYSPPPYESIDRHIKLSLD 97
F + IH AV +VGLQ I G NAR +AML +F IEDYS PP + + + ++
Sbjct 335 FTLDSIHHAVYKVGLQHLAGDISGDNARCIAMLQAFQEAIEDYSTPPMKDLTMDLTAKIN 394
Query 98 RRINFITNCRP 108
++F+ CRP
Sbjct 395 GYVSFLIECRP 405
> ath:AT2G44070 GTP binding / translation initiation factor; K03680
translation initiation factor eIF-2B subunit delta
Length=333
Score = 61.2 bits (147), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 25/66 (37%), Positives = 40/66 (60%), Gaps = 0/66 (0%)
Query 43 IHPAVIRVGLQMANRKIQGANARTVAMLSSFNCFIEDYSPPPYESIDRHIKLSLDRRINF 102
+H AV +VGL + I G NAR +AML +F ++DYS PP +++R + + ++F
Sbjct 1 MHSAVYKVGLHYLSWDISGGNARCIAMLQAFQEVVKDYSTPPENTLNRDMTAQISSYVSF 60
Query 103 ITNCRP 108
+ CRP
Sbjct 61 LIECRP 66
> sce:YGR083C GCD2, GCD12; Delta subunit of the translation initiation
factor eIF2B, the guanine-nucleotide exchange factor
for eIF2; activity subsequently regulated by phosphorylated
eIF2; first identified as a negative regulator of GCN4 expression;
K03680 translation initiation factor eIF-2B subunit
delta
Length=651
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 24/76 (31%), Positives = 44/76 (57%), Gaps = 2/76 (2%)
Query 33 RLLMNFRQEIIHPAVIRVGLQMANRKIQGANARTVAMLSSFNCFIEDYSPPPYESIDRHI 92
LL+N +++IHP+++ + +A+ KI G+ R +AML F I+DY P ++ R++
Sbjct 262 ELLLN--KDLIHPSILLLTSHLAHYKIVGSIPRCIAMLEVFQIVIKDYQTPKGTTLSRNL 319
Query 93 KLSLDRRINFITNCRP 108
L +I+ + RP
Sbjct 320 TSYLSHQIDLLKKARP 335
> bbo:BBOV_IV004670 23.m06553; hypothetical protein
Length=880
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 21/74 (28%), Positives = 34/74 (45%), Gaps = 3/74 (4%)
Query 12 STIISSSSSSNSSSSRLPLHLRLLMNFRQEIIHPAVIRVGLQMANRKIQGANARTVAMLS 71
S++I S N+S RL L++ L+ FR+ I ++R QMA ++ A+ R
Sbjct 355 SSVIRLKSVGNASVKRLQLYIASLVKFRERAISLDILR---QMATGALKDASTRVFPGTQ 411
Query 72 SFNCFIEDYSPPPY 85
F Y+ Y
Sbjct 412 QKQVFFTVYASLLY 425
Lambda K H
0.320 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2057481480
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40