bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2409_orf2
Length=89
Score E
Sequences producing significant alignments: (Bits) Value
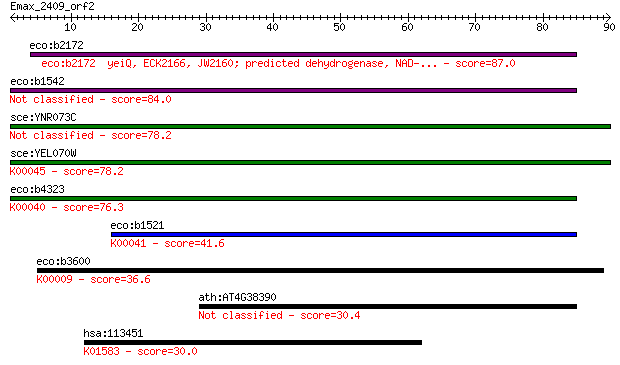
eco:b2172 yeiQ, ECK2166, JW2160; predicted dehydrogenase, NAD-... 87.0 1e-17
eco:b1542 ydfI, ECK1535, JW1535; predicted mannonate dehydroge... 84.0 1e-16
sce:YNR073C Putative mannitol dehydrogenase (EC:1.1.-.-) 78.2 6e-15
sce:YEL070W DSF1; Deletion suppressor of mpt5 mutation (EC:1.1... 78.2 6e-15
eco:b4323 uxuB, ECK4314, JW4286; D-mannonate oxidoreductase, N... 76.3 2e-14
eco:b1521 uxaB, ECK1514, JW1514; altronate oxidoreductase, NAD... 41.6 7e-04
eco:b3600 mtlD, ECK3589, JW3574; mannitol-1-phosphate dehydrog... 36.6 0.020
ath:AT4G38390 hypothetical protein 30.4 1.4
hsa:113451 ADC, AZI2, KIAA1945, ODC-p, ODC1L; arginine decarbo... 30.0 2.0
> eco:b2172 yeiQ, ECK2166, JW2160; predicted dehydrogenase, NAD-dependent;
K00540 [EC:1.-.-.-]
Length=488
Score = 87.0 bits (214), Expect = 1e-17, Method: Composition-based stats.
Identities = 37/81 (45%), Positives = 52/81 (64%), Gaps = 0/81 (0%)
Query 4 GLKLRRDNKIRPFTVLSCDNMPNNGKILKKMVIQFATEIDVEMATWISKHVCFPSTMVDR 63
LK RR+ + PFTVLSCDN+P+NG ++K V+ A + E+A WI +HV FP TMVDR
Sbjct 168 ALKRRRERGLTPFTVLSCDNIPDNGHVVKNAVLGMAEKRSPELAGWIKEHVSFPGTMVDR 227
Query 64 ITPITSKEHITLLEEDYGIKD 84
I P + E + + + G+ D
Sbjct 228 IVPAATDESLVEISQHLGVND 248
> eco:b1542 ydfI, ECK1535, JW1535; predicted mannonate dehydrogenase
Length=486
Score = 84.0 bits (206), Expect = 1e-16, Method: Composition-based stats.
Identities = 35/84 (41%), Positives = 55/84 (65%), Gaps = 0/84 (0%)
Query 1 IYMGLKLRRDNKIRPFTVLSCDNMPNNGKILKKMVIQFATEIDVEMATWISKHVCFPSTM 60
I L R+ + FTV+SCDNMP NG +++ +V +A +DV++A WI +V FPSTM
Sbjct 162 IVEALARRKAAGLPAFTVMSCDNMPENGHVMRDVVTSYAQAVDVKLAQWIEDNVTFPSTM 221
Query 61 VDRITPITSKEHITLLEEDYGIKD 84
VDRI P +++ + +E+ G++D
Sbjct 222 VDRIVPAVTEDTLAKIEQLTGVRD 245
> sce:YNR073C Putative mannitol dehydrogenase (EC:1.1.-.-)
Length=502
Score = 78.2 bits (191), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 38/90 (42%), Positives = 56/90 (62%), Gaps = 1/90 (1%)
Query 1 IYMGLKLRRDNKIRPFTVLSCDNMPNNGKILKKMVIQFAT-EIDVEMATWISKHVCFPST 59
+Y L LR + PFT++SCDNMP NG +K M++ FA + D + A WI V P++
Sbjct 177 LYEALLLRYKRGLTPFTIMSCDNMPQNGVTVKTMLVAFAKLKKDEKFAAWIEDKVTSPNS 236
Query 60 MVDRITPITSKEHITLLEEDYGIKDKWPVV 89
MVDR+TP + + + + +GIKD+ PVV
Sbjct 237 MVDRVTPRCTDKERKYVADTWGIKDQCPVV 266
> sce:YEL070W DSF1; Deletion suppressor of mpt5 mutation (EC:1.1.-.-);
K00045 mannitol 2-dehydrogenase [EC:1.1.1.67]
Length=502
Score = 78.2 bits (191), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 38/90 (42%), Positives = 56/90 (62%), Gaps = 1/90 (1%)
Query 1 IYMGLKLRRDNKIRPFTVLSCDNMPNNGKILKKMVIQFAT-EIDVEMATWISKHVCFPST 59
+Y L LR + PFT++SCDNMP NG +K M++ FA + D + A WI V P++
Sbjct 177 LYEALLLRYKRGLTPFTIMSCDNMPQNGVTVKTMLVAFAKLKKDEKFAAWIEDKVTSPNS 236
Query 60 MVDRITPITSKEHITLLEEDYGIKDKWPVV 89
MVDR+TP + + + + +GIKD+ PVV
Sbjct 237 MVDRVTPRCTDKERKYVADTWGIKDQCPVV 266
> eco:b4323 uxuB, ECK4314, JW4286; D-mannonate oxidoreductase,
NAD-binding (EC:1.1.1.57); K00040 fructuronate reductase [EC:1.1.1.57]
Length=486
Score = 76.3 bits (186), Expect = 2e-14, Method: Composition-based stats.
Identities = 34/84 (40%), Positives = 51/84 (60%), Gaps = 0/84 (0%)
Query 1 IYMGLKLRRDNKIRPFTVLSCDNMPNNGKILKKMVIQFATEIDVEMATWISKHVCFPSTM 60
I L+LRR+ ++ FTV+SCDN+ NG + K V+ A D ++A WI ++V FP TM
Sbjct 165 IVEALRLRREKGLKAFTVMSCDNVRENGHVAKVAVLGLAQARDPQLAAWIEENVTFPCTM 224
Query 61 VDRITPITSKEHITLLEEDYGIKD 84
VDRI P + E + + + G+ D
Sbjct 225 VDRIVPAATPETLQEIADQLGVYD 248
> eco:b1521 uxaB, ECK1514, JW1514; altronate oxidoreductase, NAD-dependent
(EC:1.1.1.58); K00041 tagaturonate reductase [EC:1.1.1.58]
Length=483
Score = 41.6 bits (96), Expect = 7e-04, Method: Composition-based stats.
Identities = 21/71 (29%), Positives = 40/71 (56%), Gaps = 2/71 (2%)
Query 16 FTVLSCDNMPNNGKILKKMVIQFATEIDVEMA--TWISKHVCFPSTMVDRITPITSKEHI 73
+ ++ C+ + NG L+++V+++A E + A W+ + F ST+VDRI ++ +
Sbjct 165 WIIIPCELIDYNGDALRELVLRYAQEWALPEAFIQWLDQANSFCSTLVDRIVTGYPRDEV 224
Query 74 TLLEEDYGIKD 84
LEE+ G D
Sbjct 225 AKLEEELGYHD 235
> eco:b3600 mtlD, ECK3589, JW3574; mannitol-1-phosphate dehydrogenase,
NAD(P)-binding (EC:1.1.1.17); K00009 mannitol-1-phosphate
5-dehydrogenase [EC:1.1.1.17]
Length=382
Score = 36.6 bits (83), Expect = 0.020, Method: Composition-based stats.
Identities = 22/84 (26%), Positives = 36/84 (42%), Gaps = 3/84 (3%)
Query 5 LKLRRDNKIRPFTVLSCDNMPNNGKILKKMVIQFATEIDVEMATWISKHVCFPSTMVDRI 64
+K + P +++C+NM LK V+ E + W+ +HV F + VDRI
Sbjct 104 VKRKEQGNESPLNIIACENMVRGTTQLKGHVMNALPE---DAKAWVEEHVGFVDSAVDRI 160
Query 65 TPITSKEHITLLEEDYGIKDKWPV 88
P ++ LE +W V
Sbjct 161 VPPSASATNDPLEVTVETFSEWIV 184
> ath:AT4G38390 hypothetical protein
Length=551
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 15/57 (26%), Positives = 30/57 (52%), Gaps = 1/57 (1%)
Query 29 KILKKMVIQFATEIDVEMATWISKHVCFPSTMVDRITPITSKEHITLLEE-DYGIKD 84
KI+K++ + + I + + C PS + R+ PI +K+H+ L + DY + +
Sbjct 191 KIIKELPKEEQSRISTSLQSMRVPRKCTPSCYLQRVLPILTKKHVVQLSKFDYRLSN 247
> hsa:113451 ADC, AZI2, KIAA1945, ODC-p, ODC1L; arginine decarboxylase
(EC:4.1.1.19); K01583 arginine decarboxylase [EC:4.1.1.19]
Length=460
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 12/50 (24%), Positives = 28/50 (56%), Gaps = 2/50 (4%)
Query 12 KIRPFTVLSCDNMPNNGKILKKMVIQFATEIDVEMATWISKHVCFPSTMV 61
++RPF + C++ P K+L ++ + F+ EM + +H+ P++ +
Sbjct 62 RVRPFYAVKCNSSPGVLKVLAQLGLGFSCANKAEME--LVQHIGIPASKI 109
Lambda K H
0.323 0.138 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2017039696
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40