bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2390_orf1
Length=139
Score E
Sequences producing significant alignments: (Bits) Value
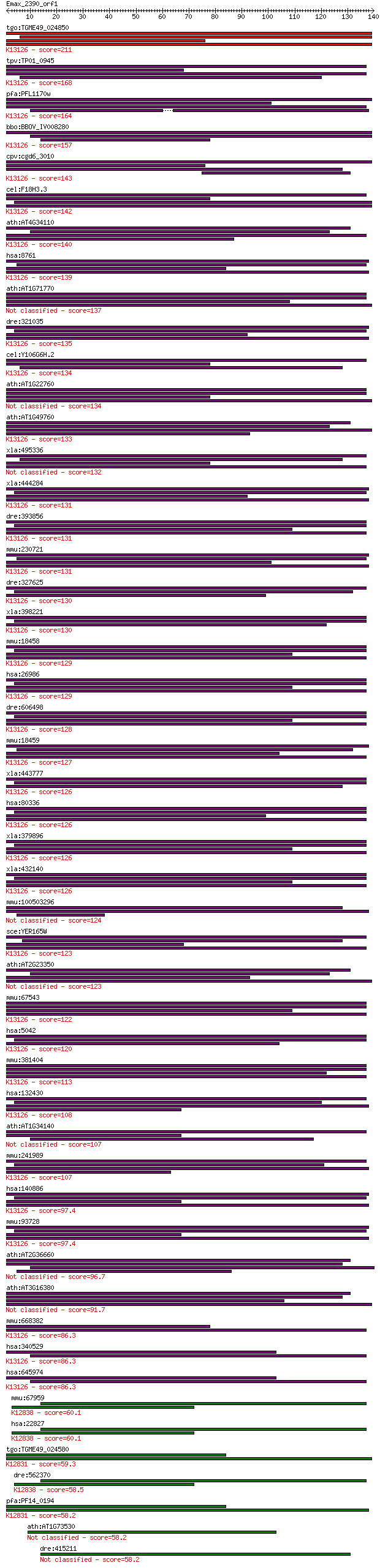
tgo:TGME49_024850 polyadenylate-binding protein, putative ; K1... 211 5e-55
tpv:TP01_0945 polyadenylate binding protein; K13126 polyadenyl... 168 4e-42
pfa:PFL1170w polyadenylate-binding protein, putative; K13126 p... 164 6e-41
bbo:BBOV_IV008280 23.m06515; polyadenylate binding protein; K1... 157 7e-39
cpv:cgd6_3010 poly(a)-binding protein fabm ; K13126 polyadenyl... 143 2e-34
cel:F18H3.3 pab-2; PolyA Binding protein family member (pab-2)... 142 4e-34
ath:AT4G34110 PAB2; PAB2 (POLY(A) BINDING 2); RNA binding / tr... 140 2e-33
hsa:8761 PABPC4, APP-1, APP1, FLJ43938, PABP4, iPABP; poly(A) ... 139 4e-33
ath:AT1G71770 PAB5; PAB5 (POLY(A)-BINDING PROTEIN 5); RNA bind... 137 1e-32
dre:321035 pabpc4, cb12, sb:cb12; poly(A) binding protein, cyt... 135 5e-32
cel:Y106G6H.2 pab-1; PolyA Binding protein family member (pab-... 134 6e-32
ath:AT1G22760 PAB3; PAB3 (POLY(A) BINDING PROTEIN 3); RNA bind... 134 1e-31
ath:AT1G49760 PAB8; PAB8 (POLY(A) BINDING PROTEIN 8); RNA bind... 133 2e-31
xla:495336 hypothetical LOC495336 132 5e-31
xla:444284 pabpc4, MGC80927, PABP, ePAB, ePABP; poly(A) bindin... 131 5e-31
dre:393856 pabpc1b, MGC77608, pabpc1, zgc:77608; poly A bindin... 131 8e-31
mmu:230721 Pabpc4, MGC11665, MGC6685; poly(A) binding protein,... 131 8e-31
dre:327625 pabpc1l, fi20g03, pab-2, wu:fi20g03, zgc:55855; pol... 130 9e-31
xla:398221 pabpc1l-a, MGC82273, ePAB, epabp-a, pabpc1l1; poly(... 130 2e-30
mmu:18458 Pabpc1, PABP, Pabp1, PabpI, Pabpl1, ePAB; poly(A) bi... 129 3e-30
hsa:26986 PABPC1, PAB1, PABP, PABP1, PABPC2, PABPL1; poly(A) b... 129 3e-30
dre:606498 pabpc1a, pabpc1, wu:fb16a02, wu:fi19b08, wu:fj12d09... 128 6e-30
mmu:18459 Pabpc2, PABP, PABP+, Pabp2; poly(A) binding protein,... 127 1e-29
xla:443777 pabpc1l-b, MGC81363, epab, epabp-b, pabpc1l1; poly(... 126 2e-29
hsa:80336 PABPC1L, C20orf119, FLJ11840, FLJ30809, FLJ42053, PA... 126 2e-29
xla:379896 pabpc1-a, MGC53109, pab1, pabp, pabp1, pabpc1, pabp... 126 2e-29
xla:432140 pabpc1-b, MGC130736, MGC79060, pab1, pabp, pabp1, p... 126 2e-29
mmu:100503296 polyadenylate-binding protein 4-like 124 8e-29
sce:YER165W PAB1; Pab1p; K13126 polyadenylate-binding protein 123 2e-28
ath:AT2G23350 PAB4; PAB4 (POLY(A) BINDING PROTEIN 4); RNA bind... 123 2e-28
mmu:67543 Pabpc6, 4932702K14Rik, AI428050, MGC132900, Pabpc3; ... 122 4e-28
hsa:5042 PABPC3, PABP3, PABPL3, tPABP; poly(A) binding protein... 120 1e-27
mmu:381404 Pabpc1l, 1810053B01Rik, Epab; poly(A) binding prote... 113 2e-25
hsa:132430 PABPC4L, DKFZp686J06116; poly(A) binding protein, c... 108 6e-24
ath:AT1G34140 PAB1; PAB1 (POLY(A) BINDING PROTEIN 1); RNA bind... 107 1e-23
mmu:241989 Pabpc4l, C330050A14Rik, EG241989; poly(A) binding p... 107 1e-23
hsa:140886 PABPC5, FLJ51677, PABP5; poly(A) binding protein, c... 97.4 1e-20
mmu:93728 Pabpc5, C820015E17, MGC130340, MGC130341; poly(A) bi... 97.4 1e-20
ath:AT2G36660 PAB7; PAB7 (POLY(A) BINDING PROTEIN 7); RNA bind... 96.7 2e-20
ath:AT3G16380 PAB6; PAB6 (POLY(A) BINDING PROTEIN 6); RNA bind... 91.7 6e-19
mmu:668382 Pabpc1l2b-ps, EG668382, Pabpc1l2b, Rbm32b; poly(A) ... 86.3 3e-17
hsa:340529 PABPC1L2A, MGC168104, PABPC1L2B, RBM32A; poly(A) bi... 86.3 3e-17
hsa:645974 PABPC1L2B, MGC168104, PABPC1L2A, RBM32B; poly(A) bi... 86.3 3e-17
mmu:67959 Puf60, 2410104I19Rik, 2810454F19Rik, SIAHBP1; poly-U... 60.1 2e-09
hsa:22827 PUF60, FIR, FLJ31379, RoBPI, SIAHBP1; poly-U binding... 60.1 2e-09
tgo:TGME49_024580 splicing factor, putative (EC:3.1.2.15); K12... 59.3 4e-09
dre:562370 puf60a, fc21a05, wu:fc21a05, wu:fi43a08; poly-U bin... 58.5 7e-09
pfa:PF14_0194 spliceosome-associated protein, putative; K12831... 58.2 7e-09
ath:AT1G73530 RNA recognition motif (RRM)-containing protein 58.2 8e-09
dre:415211 puf60b, SIAHBP1, fe37c05, repressor, si:ch211-12p8.... 58.2 8e-09
> tgo:TGME49_024850 polyadenylate-binding protein, putative ;
K13126 polyadenylate-binding protein
Length=768
Score = 211 bits (537), Expect = 5e-55, Method: Composition-based stats.
Identities = 96/138 (69%), Positives = 113/138 (81%), Gaps = 0/138 (0%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD+NIDNKALYDTFSLFGNILSCKV VD+ SKGYGFVHYE+E SAR+AI+KVNGMLIG
Sbjct 178 LDKNIDNKALYDTFSLFGNILSCKVAVDDNGHSKGYGFVHYENEESARSAIDKVNGMLIG 237
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQDP 120
GKTVYVGPFI R+ER++ ++TNVY+KN+P W D LR FG ITSLVV++DP
Sbjct 238 GKTVYVGPFIRRAERDNLAEAKYTNVYIKNMPSAWEDESRLRETFSKFGSITSLVVRKDP 297
Query 121 KGRPFAFCNYAEHSAANA 138
KGR FAFCN+A+H +A A
Sbjct 298 KGRLFAFCNFADHDSAKA 315
Score = 55.8 bits (133), Expect = 4e-08, Method: Composition-based stats.
Identities = 49/174 (28%), Positives = 73/174 (41%), Gaps = 44/174 (25%)
Query 6 DNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLI------ 59
D L +TFS FG+I S V D R + F ++ SA+ A+E +NG +
Sbjct 274 DESRLRETFSKFGSITSLVVRKDPKGRL--FAFCNFADHDSAKAAVEALNGKRVTDAGAI 331
Query 60 -------------------GGKTVYVGPFITRSERESAGVNRF-------------TNVY 87
G + ++VGP +++ R + +F N+Y
Sbjct 332 KEGEDSGAEEKEEEGQKREGDQILFVGPHQSKAHRSAMLRAKFEQMNQDRNDRFQGVNLY 391
Query 88 LKNLPRCWSDPESLRPILEPFGEITSLVVKQDPKG--RPFAF-CNYAEHSAANA 138
+KN+ D E LR + EPFG ITS V +D +G R F F C + A A
Sbjct 392 IKNMDDSIDD-EKLRQLFEPFGSITSAKVMRDERGVSRCFGFVCFMSPEEATKA 444
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 28/75 (37%), Positives = 43/75 (57%), Gaps = 0/75 (0%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
+D +ID++ L F FG+I S KV DE S+ +GFV + S A A+ +++ L+
Sbjct 395 MDDSIDDEKLRQLFEPFGSITSAKVMRDERGVSRCFGFVCFMSPEEATKAVTEMHLKLVK 454
Query 61 GKTVYVGPFITRSER 75
GK +YVG R +R
Sbjct 455 GKPLYVGLAERREQR 469
Score = 40.4 bits (93), Expect = 0.002, Method: Composition-based stats.
Identities = 35/146 (23%), Positives = 66/146 (45%), Gaps = 17/146 (11%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANR-SKGYGFVHYESEASARNAIEKVNGMLI 59
L +++ L++ F+ G + S +V D R S GY +V+Y+ A +++ +N +I
Sbjct 90 LHQDVTEAMLFEVFNSVGPVTSIRVCRDTVTRRSLGYAYVNYQGIQDAERSLDTLNYTVI 149
Query 60 GGKTVYV-----GPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSL 114
G+ + P + +S N+++KNL + D ++L FG I S
Sbjct 150 KGQPCRIMWCHRDPSLRKSGN--------GNIFVKNLDKN-IDNKALYDTFSLFGNILSC 200
Query 115 VVKQDPKG--RPFAFCNYAEHSAANA 138
V D G + + F +Y +A +
Sbjct 201 KVAVDDNGHSKGYGFVHYENEESARS 226
> tpv:TP01_0945 polyadenylate binding protein; K13126 polyadenylate-binding
protein
Length=661
Score = 168 bits (426), Expect = 4e-42, Method: Compositional matrix adjust.
Identities = 85/136 (62%), Positives = 102/136 (75%), Gaps = 3/136 (2%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD++ID K+LYDTFS FG ILSCKV VD + SK YGFVHYE+E SAR AIEKVNGMLIG
Sbjct 122 LDKSIDTKSLYDTFSHFGPILSCKVAVDASGASKRYGFVHYENEESAREAIEKVNGMLIG 181
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQDP 120
GK V V PF+ + +RES V FTN+Y++N P W D E+LR LE +GEITS+++K+D
Sbjct 182 GKRVEVAPFLRKQDRESEEV--FTNLYVRNFPADW-DEEALRQFLEKYGEITSMMLKEDS 238
Query 121 KGRPFAFCNYAEHSAA 136
KGR FAF NY E A
Sbjct 239 KGRRFAFVNYKEPEVA 254
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 27/67 (40%), Positives = 41/67 (61%), Gaps = 0/67 (0%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD + D+++L + F FG I S KV +D N S+G+GFV + + A AI ++ L+
Sbjct 313 LDDSFDDESLGELFKPFGTITSSKVMLDANNHSRGFGFVCFTNPQEATKAIAAMHLKLVK 372
Query 61 GKTVYVG 67
GK +YVG
Sbjct 373 GKPLYVG 379
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 43/144 (29%), Positives = 66/144 (45%), Gaps = 17/144 (11%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANR-SKGYGFVHYESEASARNAIEKVNGMLI 59
L ++ LY+ F+ G + S +V D R S GY +V+Y S A+ A+E +N + I
Sbjct 34 LKPDVTEAVLYEVFNTVGPVASIRVCRDSVTRKSLGYAYVNYYSTQDAQEALENLNYIEI 93
Query 60 GGKTVYV-----GPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSL 114
G + P + RS AG N+++KNL + D +SL FG I S
Sbjct 94 KGHPTRIMWSNRDPSLRRS---GAG-----NIFVKNLDKSI-DTKSLYDTFSHFGPILSC 144
Query 115 VVKQDPKG--RPFAFCNYAEHSAA 136
V D G + + F +Y +A
Sbjct 145 KVAVDASGASKRYGFVHYENEESA 168
Score = 42.0 bits (97), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 34/129 (26%), Positives = 59/129 (45%), Gaps = 18/129 (13%)
Query 6 DNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLI--GGKT 63
D +AL +G I S + D R + FV+Y+ A+ + +N + + +
Sbjct 215 DEEALRQFLEKYGEITSMMLKEDSKGRR--FAFVNYKEPEVAKEVVNTLNDLKLEESSEP 272
Query 64 VYVGPFITRSERESAGVNRF-------------TNVYLKNLPRCWSDPESLRPILEPFGE 110
+ V P +++R++ +F +N+Y+KNL + D ESL + +PFG
Sbjct 273 LLVCPHQDKAKRQNLLRAQFNNSTMAQEDKRVTSNLYIKNLDDSFDD-ESLGELFKPFGT 331
Query 111 ITSLVVKQD 119
ITS V D
Sbjct 332 ITSSKVMLD 340
> pfa:PFL1170w polyadenylate-binding protein, putative; K13126
polyadenylate-binding protein
Length=875
Score = 164 bits (416), Expect = 6e-41, Method: Composition-based stats.
Identities = 83/139 (59%), Positives = 103/139 (74%), Gaps = 3/139 (2%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD++IDNKAL+DTFS+FGNILSCKV DE +SK YGFVHYE E SA+ AIEKVNG+ +G
Sbjct 111 LDKSIDNKALFDTFSMFGNILSCKVATDEFGKSKSYGFVHYEDEESAKEAIEKVNGVQLG 170
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQDP 120
K VYVGPFI +SER + +FTN+Y+KN P ++ LR + P+GEITS++VK D
Sbjct 171 SKNVYVGPFIKKSERATND-TKFTNLYVKNFPDSVTETH-LRQLFNPYGEITSMIVKMDN 228
Query 121 KGRPFAFCNYAE-HSAANA 138
K R F F NYA+ SA NA
Sbjct 229 KNRKFCFINYADAESAKNA 247
Score = 59.3 bits (142), Expect = 4e-09, Method: Composition-based stats.
Identities = 38/107 (35%), Positives = 56/107 (52%), Gaps = 7/107 (6%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD ID+ L + F FG I S KV DE +SKG+GFV + S+ A A+ +++ +I
Sbjct 457 LDDGIDDIMLRELFEPFGTITSAKVMRDEKEQSKGFGFVCFASQEEANKAVTEMHLKIIN 516
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLK-------NLPRCWSDPES 100
GK +YVG R +R S RF ++ N P ++ P+S
Sbjct 517 GKPLYVGLAEKREQRLSRLQQRFRMHPIRHHMNNPLNTPMQYASPQS 563
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 39/140 (27%), Positives = 68/140 (48%), Gaps = 9/140 (6%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANR-SKGYGFVHYESEASARNAIEKVNGMLI 59
L+ ++ LY+ F+ G++ S +V D R S GY +V+Y + A A A++ +N I
Sbjct 23 LNEDVTEAVLYEIFNTVGHVSSIRVCRDSVTRKSLGYAYVNYHNLADAERALDTLNYTNI 82
Query 60 GGKTVYVGPFITRSERESAGVNRFT-NVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQ 118
G+ + S R+ + T N+++KNL + D ++L FG I S V
Sbjct 83 KGQPAR----LMWSHRDPSLRKSGTGNIFVKNLDKS-IDNKALFDTFSMFGNILSCKVAT 137
Query 119 DP--KGRPFAFCNYAEHSAA 136
D K + + F +Y + +A
Sbjct 138 DEFGKSKSYGFVHYEDEESA 157
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 30/89 (33%), Positives = 42/89 (47%), Gaps = 16/89 (17%)
Query 64 VYVGPFITRSER-------------ESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGE 110
+YVGP +R+ R E+ ++ N+Y+KNL D LR + EPFG
Sbjct 417 LYVGPHQSRARRHAILKAKFDNLNVENKNKHQGVNLYIKNLDDGIDDI-MLRELFEPFGT 475
Query 111 ITSLVVKQDPK--GRPFAFCNYAEHSAAN 137
ITS V +D K + F F +A AN
Sbjct 476 ITSAKVMRDEKEQSKGFGFVCFASQEEAN 504
Score = 32.7 bits (73), Expect = 0.39, Method: Composition-based stats.
Identities = 18/50 (36%), Positives = 28/50 (56%), Gaps = 2/50 (4%)
Query 10 LYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLI 59
L F+ +G I S V +D NR + F++Y SA+NA++ +NG I
Sbjct 209 LRQLFNPYGEITSMIVKMDNKNRK--FCFINYADAESAKNAMDNLNGKKI 256
> bbo:BBOV_IV008280 23.m06515; polyadenylate binding protein;
K13126 polyadenylate-binding protein
Length=585
Score = 157 bits (398), Expect = 7e-39, Method: Composition-based stats.
Identities = 77/138 (55%), Positives = 96/138 (69%), Gaps = 1/138 (0%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD+ ID KALYDTFS FG ILSCKV +D SKGYGFVHY +E SA+ AIEKVNGMLIG
Sbjct 131 LDKAIDTKALYDTFSHFGTILSCKVAIDSLGNSKGYGFVHYTTEESAKEAIEKVNGMLIG 190
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQDP 120
V V PF+ R+ER S + FTN+Y++N P W++ + L +GEITSL++K D
Sbjct 191 NSQVSVAPFLRRNERTSTVGDVFTNLYVRNFPDTWTE-DDLHQTFSKYGEITSLLLKSDD 249
Query 121 KGRPFAFCNYAEHSAANA 138
KGR FAF N+ + + A A
Sbjct 250 KGRRFAFVNFVDTNMAKA 267
Score = 42.7 bits (99), Expect = 3e-04, Method: Composition-based stats.
Identities = 43/134 (32%), Positives = 65/134 (48%), Gaps = 10/134 (7%)
Query 10 LYDTFSLFGNILSCKVGVDEANR-SKGYGFVHYESEASARNAIEKVNGMLIGGKTVYVGP 68
LY+ F+ G + S +V D R S GY +V+Y + A A+E +N + I G+
Sbjct 52 LYEVFNGIGPVASIRVCRDSLTRKSLGYAYVNYYNFQDAEAALECLNYIEIKGQPAR--- 108
Query 69 FITRSERESAGVNRFT-NVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQDPKG--RPF 125
I SER+ + T N+++KNL + D ++L FG I S V D G + +
Sbjct 109 -IMWSERDPSLRKSGTGNIFVKNLDKA-IDTKALYDTFSHFGTILSCKVAIDSLGNSKGY 166
Query 126 AFCNY-AEHSAANA 138
F +Y E SA A
Sbjct 167 GFVHYTTEESAKEA 180
Score = 37.7 bits (86), Expect = 0.010, Method: Composition-based stats.
Identities = 21/64 (32%), Positives = 32/64 (50%), Gaps = 0/64 (0%)
Query 14 FSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKTVYVGPFITRS 73
F +G + S KV D S+G+GFV + A A+ ++ L+ K +YVG R
Sbjct 337 FKQYGTVTSSKVMRDHNGVSRGFGFVCFSRPDEATKAVAGMHLKLVKNKPLYVGLAEKRE 396
Query 74 ERES 77
+R S
Sbjct 397 QRAS 400
> cpv:cgd6_3010 poly(a)-binding protein fabm ; K13126 polyadenylate-binding
protein
Length=746
Score = 143 bits (360), Expect = 2e-34, Method: Composition-based stats.
Identities = 66/138 (47%), Positives = 97/138 (70%), Gaps = 1/138 (0%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD++IDNK L+DTFSLFGNI+SCK+ D +S GYGF+H+E SA+ AI ++NG ++G
Sbjct 107 LDKSIDNKTLFDTFSLFGNIMSCKIATDVEGKSLGYGFIHFEHADSAKEAISRLNGAVLG 166
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQDP 120
+ +YVG F ++ER S FTNVY+K++P+ W++ + L I +G+I+SLV++ D
Sbjct 167 DRPIYVGKFQKKAERFSEKDKTFTNVYVKHIPKSWTE-DLLYKIFGVYGKISSLVLQSDS 225
Query 121 KGRPFAFCNYAEHSAANA 138
KGRPF F N+ +A A
Sbjct 226 KGRPFGFVNFENPDSAKA 243
Score = 45.8 bits (107), Expect = 4e-05, Method: Composition-based stats.
Identities = 25/75 (33%), Positives = 41/75 (54%), Gaps = 0/75 (0%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
L +I+ + L F FG + S + DE+ S+G+GFV + S A AI +++ L+
Sbjct 370 LADSINEEDLRSMFEPFGTVSSVSIKTDESGVSRGFGFVSFLSPDEATKAITEMHLKLVR 429
Query 61 GKTVYVGPFITRSER 75
GK +YVG + +R
Sbjct 430 GKPLYVGLHERKEQR 444
Score = 43.9 bits (102), Expect = 2e-04, Method: Composition-based stats.
Identities = 37/129 (28%), Positives = 60/129 (46%), Gaps = 7/129 (5%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDE-ANRSKGYGFVHYESEASARNAIEKVNGMLI 59
LD ++ LY+ F+ + S ++ D RS GY +V+Y S A A A++ +N I
Sbjct 19 LDADVTETMLYEIFNSVAVVSSVRICRDALTRRSLGYAYVNYNSVADAERALDTLNFTCI 78
Query 60 GGKTVYVGPFITRSERESAG-VNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQ 118
G+ I R+ A N NV++KNL + D ++L FG I S +
Sbjct 79 RGRPCR----IMWCLRDPASRRNNDGNVFVKNLDKS-IDNKTLFDTFSLFGNIMSCKIAT 133
Query 119 DPKGRPFAF 127
D +G+ +
Sbjct 134 DVEGKSLGY 142
Score = 37.4 bits (85), Expect = 0.014, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 34/58 (58%), Gaps = 3/58 (5%)
Query 75 RESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQDPKG--RPFAFCNY 130
+ES + N+Y+KNL ++ E LR + EPFG ++S+ +K D G R F F ++
Sbjct 354 KESHQRYQGVNLYVKNLADSINE-EDLRSMFEPFGTVSSVSIKTDESGVSRGFGFVSF 410
> cel:F18H3.3 pab-2; PolyA Binding protein family member (pab-2);
K13126 polyadenylate-binding protein
Length=566
Score = 142 bits (357), Expect = 4e-34, Method: Composition-based stats.
Identities = 77/142 (54%), Positives = 97/142 (68%), Gaps = 8/142 (5%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LDR IDNK++YDTFSLFGNILSCKV D+ SKGYGFVH+E+E SA+ AIEKVNGML+
Sbjct 152 LDRVIDNKSVYDTFSLFGNILSCKVATDDEGNSKGYGFVHFETEHSAQTAIEKVNGMLLS 211
Query 61 GKTVYVGPFITRSERE----SAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVV 116
K VYVG F R++R +G+ ++TNV++KN D E L + FGEITS VV
Sbjct 212 DKKVYVGKFQPRAQRMKELGESGL-KYTNVFVKNFGEHL-DQEKLSAMFSKFGEITSAVV 269
Query 117 KQDPKGRP--FAFCNYAEHSAA 136
D +G+P F F +A+ AA
Sbjct 270 MTDAQGKPKGFGFVAFADQDAA 291
Score = 60.1 bits (144), Expect = 2e-09, Method: Composition-based stats.
Identities = 30/77 (38%), Positives = 45/77 (58%), Gaps = 0/77 (0%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
++ ++++ L D FS FG I S KV VDE RSKG+GFV +E A A+ +N +IG
Sbjct 350 IEEDLNDDGLRDHFSSFGTITSAKVMVDENGRSKGFGFVCFEKPEEATAAVTDMNSKMIG 409
Query 61 GKTVYVGPFITRSERES 77
K +YV + +R +
Sbjct 410 AKPLYVALAQRKEDRRA 426
Score = 55.8 bits (133), Expect = 4e-08, Method: Composition-based stats.
Identities = 45/154 (29%), Positives = 71/154 (46%), Gaps = 22/154 (14%)
Query 4 NIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKT 63
++D + L FS FG I S V D + KG+GFV + + +A A+EK+N ++ G
Sbjct 248 HLDQEKLSAMFSKFGEITSAVVMTDAQGKPKGFGFVAFADQDAAGQAVEKLNDSILEGTD 307
Query 64 VYVGPFITRSERES---------------AGVNRF--TNVYLKNLPRCWSDPESLRPILE 106
+ + R++++S V R+ N+Y+KN+ +D + LR
Sbjct 308 CKLS--VCRAQKKSERSAELKRKYEALKQERVQRYQGVNLYVKNIEEDLND-DGLRDHFS 364
Query 107 PFGEITSLVVKQDPKGRP--FAFCNYAEHSAANA 138
FG ITS V D GR F F + + A A
Sbjct 365 SFGTITSAKVMVDENGRSKGFGFVCFEKPEEATA 398
Score = 54.7 bits (130), Expect = 8e-08, Method: Composition-based stats.
Identities = 44/143 (30%), Positives = 74/143 (51%), Gaps = 10/143 (6%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANR-SKGYGFVHYESEASARNAIEKVNGMLI 59
L ++ L++ FS+ G +LS +V D +R S GY +V+++ A A A++ +N +I
Sbjct 64 LHPDVSEAMLFEKFSMAGPVLSIRVCRDNTSRLSLGYAYVNFQQPADAERALDTMNFEVI 123
Query 60 GGKTVYVGPFITRSERESAGVNRFT-NVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQ 118
G+ + I S+R+ A N+++KNL R D +S+ FG I S V
Sbjct 124 HGRPMR----IMWSQRDPAARRAGNGNIFIKNLDRV-IDNKSVYDTFSLFGNILSCKVAT 178
Query 119 DPKG--RPFAFCNY-AEHSAANA 138
D +G + + F ++ EHSA A
Sbjct 179 DDEGNSKGYGFVHFETEHSAQTA 201
> ath:AT4G34110 PAB2; PAB2 (POLY(A) BINDING 2); RNA binding /
translation initiation factor; K13126 polyadenylate-binding
protein
Length=629
Score = 140 bits (352), Expect = 2e-33, Method: Composition-based stats.
Identities = 73/133 (54%), Positives = 96/133 (72%), Gaps = 4/133 (3%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD +ID+KAL+DTFS FGNI+SCKV VD + +SKGYGFV Y +E SA+ AIEK+NGML+
Sbjct 131 LDESIDHKALHDTFSSFGNIVSCKVAVDSSGQSKGYGFVQYANEESAQKAIEKLNGMLLN 190
Query 61 GKTVYVGPFITRSERES-AGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQD 119
K VYVGPF+ R ER+S A +FTNVY+KNL +D + L+ +G+ITS VV +D
Sbjct 191 DKQVYVGPFLRRQERDSTANKTKFTNVYVKNLAESTTD-DDLKNAFGEYGKITSAVVMKD 249
Query 120 PKGRP--FAFCNY 130
+G+ F F N+
Sbjct 250 GEGKSKGFGFVNF 262
Score = 72.8 bits (177), Expect = 3e-13, Method: Composition-based stats.
Identities = 47/126 (37%), Positives = 65/126 (51%), Gaps = 14/126 (11%)
Query 10 LYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKTVYVGPF 69
L + F +G I S V D +SKG+GFV++E+ A A+E +NG K YVG
Sbjct 231 LKNAFGEYGKITSAVVMKDGEGKSKGFGFVNFENADDAARAVESLNGHKFDDKEWYVGRA 290
Query 70 ITRSERESAGVNRF-------------TNVYLKNLPRCWSDPESLRPILEPFGEITSLVV 116
+SERE+ R+ +N+Y+KNL SD E L+ I PFG +TS V
Sbjct 291 QKKSERETELRVRYEQNLKEAADKFQSSNLYVKNLDPSISD-EKLKEIFSPFGTVTSSKV 349
Query 117 KQDPKG 122
+DP G
Sbjct 350 MRDPNG 355
Score = 56.2 bits (134), Expect = 3e-08, Method: Composition-based stats.
Identities = 44/144 (30%), Positives = 70/144 (48%), Gaps = 17/144 (11%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVD-EANRSKGYGFVHYESEASARNAIEKVNGMLI 59
LD N+ + L+D F G +++ +V D RS GYG+V++ + A AI+++N + +
Sbjct 43 LDFNVTDSQLFDAFGQMGTVVTVRVCRDLVTRRSLGYGYVNFTNPQDAARAIQELNYIPL 102
Query 60 GGKTVYV-----GPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSL 114
GK + V P + RS AG N+++KNL D ++L FG I S
Sbjct 103 YGKPIRVMYSHRDPSVRRS---GAG-----NIFIKNLDES-IDHKALHDTFSSFGNIVSC 153
Query 115 VVKQDPKGRP--FAFCNYAEHSAA 136
V D G+ + F YA +A
Sbjct 154 KVAVDSSGQSKGYGFVQYANEESA 177
Score = 47.8 bits (112), Expect = 1e-05, Method: Composition-based stats.
Identities = 28/86 (32%), Positives = 45/86 (52%), Gaps = 0/86 (0%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD +I ++ L + FS FG + S KV D SKG GFV + + A A+ +++G +I
Sbjct 325 LDPSISDEKLKEIFSPFGTVTSSKVMRDPNGTSKGSGFVAFATPEEATEAMSQLSGKMIE 384
Query 61 GKTVYVGPFITRSERESAGVNRFTNV 86
K +YV + +R +F+ V
Sbjct 385 SKPLYVAIAQRKEDRRVRLQAQFSQV 410
> hsa:8761 PABPC4, APP-1, APP1, FLJ43938, PABP4, iPABP; poly(A)
binding protein, cytoplasmic 4 (inducible form); K13126 polyadenylate-binding
protein
Length=660
Score = 139 bits (349), Expect = 4e-33, Method: Composition-based stats.
Identities = 75/142 (52%), Positives = 96/142 (67%), Gaps = 7/142 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD++IDNKALYDTFS FGNILSCKV DE N SKGY FVH+E++ +A AIEK+NGML+
Sbjct 106 LDKSIDNKALYDTFSAFGNILSCKVVCDE-NGSKGYAFVHFETQEAADKAIEKMNGMLLN 164
Query 61 GKTVYVGPFITRSERES---AGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK 117
+ V+VG F +R ERE+ A FTNVY+KN D ESL+ + FG+ S+ V
Sbjct 165 DRKVFVGRFKSRKEREAELGAKAKEFTNVYIKNFGE-EVDDESLKELFSQFGKTLSVKVM 223
Query 118 QDPKGRP--FAFCNYAEHSAAN 137
+DP G+ F F +Y +H AN
Sbjct 224 RDPNGKSKGFGFVSYEKHEDAN 245
Score = 83.2 bits (204), Expect = 2e-16, Method: Composition-based stats.
Identities = 52/146 (35%), Positives = 76/146 (52%), Gaps = 15/146 (10%)
Query 5 IDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKTV 64
+D+++L + FS FG LS KV D +SKG+GFV YE A A+E++NG I GK +
Sbjct 202 VDDESLKELFSQFGKTLSVKVMRDPNGKSKGFGFVSYEKHEDANKAVEEMNGKEISGKII 261
Query 65 YVGPFITRSERESAGVNRF-------------TNVYLKNLPRCWSDPESLRPILEPFGEI 111
+VG + ER++ +F N+Y+KNL D E LR PFG I
Sbjct 262 FVGRAQKKVERQAELKRKFEQLKQERISRYQGVNLYIKNLDDTIDD-EKLRKEFSPFGSI 320
Query 112 TSL-VVKQDPKGRPFAFCNYAEHSAA 136
TS V+ +D + + F F ++ A
Sbjct 321 TSAKVMLEDGRSKGFGFVCFSSPEEA 346
Score = 60.5 bits (145), Expect = 2e-09, Method: Composition-based stats.
Identities = 32/83 (38%), Positives = 51/83 (61%), Gaps = 1/83 (1%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD ID++ L FS FG+I S KV +++ RSKG+GFV + S A A+ ++NG ++G
Sbjct 301 LDDTIDDEKLRKEFSPFGSITSAKVMLEDG-RSKGFGFVCFSSPEEATKAVTEMNGRIVG 359
Query 61 GKTVYVGPFITRSERESAGVNRF 83
K +YV + ER++ N++
Sbjct 360 SKPLYVALAQRKEERKAHLTNQY 382
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 45/143 (31%), Positives = 72/143 (50%), Gaps = 14/143 (9%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDE-ANRSKGYGFVHYESEASARNAIEKVNGMLI 59
L ++ LY+ FS G +LS +V D RS GY +V+++ A A A++ +N +I
Sbjct 18 LHSDVTEAMLYEKFSPAGPVLSIRVCRDMITRRSLGYAYVNFQQPADAERALDTMNFDVI 77
Query 60 GGKTVYVGPFITRSERE----SAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLV 115
GK + I S+R+ +GV NV++KNL + D ++L FG I S
Sbjct 78 KGKPIR----IMWSQRDPSLRKSGVG---NVFIKNLDKS-IDNKALYDTFSAFGNILSCK 129
Query 116 VKQDPKG-RPFAFCNYAEHSAAN 137
V D G + +AF ++ AA+
Sbjct 130 VVCDENGSKGYAFVHFETQEAAD 152
> ath:AT1G71770 PAB5; PAB5 (POLY(A)-BINDING PROTEIN 5); RNA binding
/ poly(A) binding / translation initiation factor
Length=668
Score = 137 bits (344), Expect = 1e-32, Method: Composition-based stats.
Identities = 74/141 (52%), Positives = 95/141 (67%), Gaps = 6/141 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD +IDNKALY+TFS FG ILSCKV +D RSKGYGFV +E E +A+ AI+K+NGML+
Sbjct 139 LDASIDNKALYETFSSFGTILSCKVAMDVVGRSKGYGFVQFEKEETAQAAIDKLNGMLLN 198
Query 61 GKTVYVGPFITRSER---ESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK 117
K V+VG F+ R +R ES V FTNVY+KNLP+ +D E L+ +G+I+S VV
Sbjct 199 DKQVFVGHFVRRQDRARSESGAVPSFTNVYVKNLPKEITDDE-LKKTFGKYGDISSAVVM 257
Query 118 QDPKG--RPFAFCNYAEHSAA 136
+D G R F F N+ AA
Sbjct 258 KDQSGNSRSFGFVNFVSPEAA 278
Score = 70.9 bits (172), Expect = 1e-12, Method: Composition-based stats.
Identities = 49/151 (32%), Positives = 79/151 (52%), Gaps = 16/151 (10%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
L + I + L TF +G+I S V D++ S+ +GFV++ S +A A+EK+NG+ +G
Sbjct 232 LPKEITDDELKKTFGKYGDISSAVVMKDQSGNSRSFGFVNFVSPEAAAVAVEKMNGISLG 291
Query 61 GKTVYVGPFITRSER--------ESAGVNRF-----TNVYLKNLPRCWSDPESLRPILEP 107
+YVG +S+R E ++RF +N+YLKNL +D E L+ +
Sbjct 292 EDVLYVGRAQKKSDREEELRRKFEQERISRFEKLQGSNLYLKNLDDSVND-EKLKEMFSE 350
Query 108 FGEITSLVVKQDPKG--RPFAFCNYAEHSAA 136
+G +TS V + +G R F F Y+ A
Sbjct 351 YGNVTSCKVMMNSQGLSRGFGFVAYSNPEEA 381
Score = 62.0 bits (149), Expect = 6e-10, Method: Composition-based stats.
Identities = 33/107 (30%), Positives = 60/107 (56%), Gaps = 9/107 (8%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD +++++ L + FS +GN+ SCKV ++ S+G+GFV Y + A A++++NG +IG
Sbjct 335 LDDSVNDEKLKEMFSEYGNVTSCKVMMNSQGLSRGFGFVAYSNPEEALLAMKEMNGKMIG 394
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEP 107
K +YV + ER++ + FT + P ++ P+ P
Sbjct 395 RKPLYVALAQRKEERQAHLQSLFTQI---------RSPGTMSPVPSP 432
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 36/140 (25%), Positives = 64/140 (45%), Gaps = 6/140 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD +++ L D F+ + + +V D +RS GY +V++ + A A+E +N I
Sbjct 52 LDPSVNESHLLDLFNQVAPVHNLRVCRDLTHRSLGYAYVNFANPEDASRAMESLNYAPIR 111
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQDP 120
+ + + + + S ++ NV++KNL D ++L FG I S V D
Sbjct 112 DRPIRI---MLSNRDPSTRLSGKGNVFIKNLDAS-IDNKALYETFSSFGTILSCKVAMDV 167
Query 121 KGRP--FAFCNYAEHSAANA 138
GR + F + + A A
Sbjct 168 VGRSKGYGFVQFEKEETAQA 187
> dre:321035 pabpc4, cb12, sb:cb12; poly(A) binding protein, cytoplasmic
4 (inducible form); K13126 polyadenylate-binding
protein
Length=637
Score = 135 bits (339), Expect = 5e-32, Method: Composition-based stats.
Identities = 73/142 (51%), Positives = 95/142 (66%), Gaps = 7/142 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD++IDNKALYDTFS FGNILSCKV DE N SKGY FVH+E++ +A AIEK+NGML+
Sbjct 107 LDKSIDNKALYDTFSAFGNILSCKVVCDE-NGSKGYAFVHFETQDAADRAIEKMNGMLLN 165
Query 61 GKTVYVGPFITRSERES---AGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK 117
+ V+VG F +R ERE+ A FTNVY+KN D + L+ + + +G+ S+ V
Sbjct 166 DRKVFVGRFKSRKEREAEMGAKAKEFTNVYIKNFGDDMDD-QRLKELFDKYGKTLSVKVM 224
Query 118 QDPKG--RPFAFCNYAEHSAAN 137
DP G R F F +Y +H AN
Sbjct 225 TDPTGKSRGFGFVSYEKHEDAN 246
Score = 80.5 bits (197), Expect = 1e-15, Method: Composition-based stats.
Identities = 49/147 (33%), Positives = 76/147 (51%), Gaps = 15/147 (10%)
Query 4 NIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKT 63
++D++ L + F +G LS KV D +S+G+GFV YE A A+E++NG + GKT
Sbjct 202 DMDDQRLKELFDKYGKTLSVKVMTDPTGKSRGFGFVSYEKHEDANKAVEEMNGTELNGKT 261
Query 64 VYVGPFITRSERESAGVNRF-------------TNVYLKNLPRCWSDPESLRPILEPFGE 110
V+VG + ER++ +F N+Y+KNL D E LR PFG
Sbjct 262 VFVGRAQKKMERQAELKRKFEQLKQERISRYQGVNLYIKNLDDTIDD-EKLRKEFSPFGS 320
Query 111 ITSL-VVKQDPKGRPFAFCNYAEHSAA 136
ITS V+ ++ + + F F ++ A
Sbjct 321 ITSAKVMLEEGRSKGFGFVCFSSPEEA 347
Score = 62.4 bits (150), Expect = 4e-10, Method: Composition-based stats.
Identities = 35/91 (38%), Positives = 54/91 (59%), Gaps = 5/91 (5%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD ID++ L FS FG+I S KV ++E RSKG+GFV + S A A+ ++NG ++G
Sbjct 302 LDDTIDDEKLRKEFSPFGSITSAKVMLEEG-RSKGFGFVCFSSPEEATKAVTEMNGRIVG 360
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNL 91
K +YV + ER++ TN Y++ +
Sbjct 361 SKPLYVALAQRKEERKA----HLTNQYMQRI 387
Score = 52.8 bits (125), Expect = 4e-07, Method: Composition-based stats.
Identities = 45/143 (31%), Positives = 72/143 (50%), Gaps = 14/143 (9%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDE-ANRSKGYGFVHYESEASARNAIEKVNGMLI 59
L +I LY+ FS G +LS +V D RS GY +V+++ A A A++ +N ++
Sbjct 19 LHPDITEAMLYEKFSPAGPVLSIRVCRDMITRRSLGYAYVNFQQPADAERALDTMNFDVV 78
Query 60 GGKTVYVGPFITRSERE----SAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLV 115
GK + I S+R+ +GV NV++KNL + D ++L FG I S
Sbjct 79 KGKPIR----IMWSQRDPSLRKSGVG---NVFIKNLDKS-IDNKALYDTFSAFGNILSCK 130
Query 116 VKQDPKG-RPFAFCNYAEHSAAN 137
V D G + +AF ++ AA+
Sbjct 131 VVCDENGSKGYAFVHFETQDAAD 153
> cel:Y106G6H.2 pab-1; PolyA Binding protein family member (pab-1);
K13126 polyadenylate-binding protein
Length=583
Score = 134 bits (338), Expect = 6e-32, Method: Composition-based stats.
Identities = 71/140 (50%), Positives = 94/140 (67%), Gaps = 5/140 (3%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD+ IDNK++YDTFSLFGNILSCKV +DE SKGYGFVH+E+E +A+NAI+KVNGML+
Sbjct 64 LDKVIDNKSIYDTFSLFGNILSCKVAIDEDGFSKGYGFVHFETEEAAQNAIQKVNGMLLA 123
Query 61 GKTVYVGPFITRSERE---SAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSL-VV 116
GK V+VG F R++R +FTNVY+KN + + E+L + FG ITS V+
Sbjct 124 GKKVFVGKFQPRAQRNRELGETAKQFTNVYVKNFGDHY-NKETLEKLFAKFGNITSCEVM 182
Query 117 KQDPKGRPFAFCNYAEHSAA 136
+ K + F F +A A
Sbjct 183 TVEGKSKGFGFVAFANPEEA 202
Score = 58.2 bits (139), Expect = 8e-09, Method: Composition-based stats.
Identities = 28/77 (36%), Positives = 43/77 (55%), Gaps = 0/77 (0%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD +D+ L F +GNI S KV DE RSKG+GFV +E A +A+ ++N ++
Sbjct 261 LDETVDDDGLKKQFESYGNITSAKVMTDENGRSKGFGFVCFEKPEEATSAVTEMNSKMVC 320
Query 61 GKTVYVGPFITRSERES 77
K +YV + +R +
Sbjct 321 SKPLYVAIAQRKEDRRA 337
Score = 52.0 bits (123), Expect = 5e-07, Method: Composition-based stats.
Identities = 43/137 (31%), Positives = 63/137 (45%), Gaps = 17/137 (12%)
Query 6 DNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGK--T 63
+ + L F+ FGNI SC+V E +SKG+GFV + + A A++ ++ I G
Sbjct 162 NKETLEKLFAKFGNITSCEVMTVEG-KSKGFGFVAFANPEEAETAVQALHDSTIEGTDLK 220
Query 64 VYVGPFITRSER--------ESAGVNRF-----TNVYLKNLPRCWSDPESLRPILEPFGE 110
++V +SER E R N+Y+KNL D + L+ E +G
Sbjct 221 LHVCRAQKKSERHAELKKKHEQHKAERMQKYQGVNLYVKNLDETVDD-DGLKKQFESYGN 279
Query 111 ITSLVVKQDPKGRPFAF 127
ITS V D GR F
Sbjct 280 ITSAKVMTDENGRSKGF 296
> ath:AT1G22760 PAB3; PAB3 (POLY(A) BINDING PROTEIN 3); RNA binding
/ translation initiation factor
Length=660
Score = 134 bits (336), Expect = 1e-31, Method: Composition-based stats.
Identities = 76/141 (53%), Positives = 93/141 (65%), Gaps = 6/141 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD +IDNKAL++TFS FG ILSCKV +D RSKGYGFV +E E SA+ AI+K+NGML+
Sbjct 143 LDASIDNKALFETFSSFGTILSCKVAMDVTGRSKGYGFVQFEKEESAQAAIDKLNGMLMN 202
Query 61 GKTVYVGPFITRSER---ESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK 117
K V+VG FI R ER E+ RFTNVY+KNLP+ + E LR FG I+S VV
Sbjct 203 DKQVFVGHFIRRQERARDENTPTPRFTNVYVKNLPKEIGEDE-LRKTFGKFGVISSAVVM 261
Query 118 QDPKG--RPFAFCNYAEHSAA 136
+D G R F F N+ AA
Sbjct 262 RDQSGNSRCFGFVNFECTEAA 282
Score = 77.0 bits (188), Expect = 2e-14, Method: Composition-based stats.
Identities = 53/151 (35%), Positives = 77/151 (50%), Gaps = 16/151 (10%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
L + I L TF FG I S V D++ S+ +GFV++E +A +A+EK+NG+ +G
Sbjct 236 LPKEIGEDELRKTFGKFGVISSAVVMRDQSGNSRCFGFVNFECTEAAASAVEKMNGISLG 295
Query 61 GKTVYVGPFITRSER--------ESAGVNRF-----TNVYLKNLPRCWSDPESLRPILEP 107
+YVG +SER E +NRF N+YLKNL D E L+ +
Sbjct 296 DDVLYVGRAQKKSEREEELRRKFEQERINRFEKSQGANLYLKNLDDSVDD-EKLKEMFSE 354
Query 108 FGEITSLVVKQDPKG--RPFAFCNYAEHSAA 136
+G +TS V +P+G R F F Y+ A
Sbjct 355 YGNVTSSKVMLNPQGMSRGFGFVAYSNPEEA 385
Score = 55.8 bits (133), Expect = 4e-08, Method: Composition-based stats.
Identities = 26/77 (33%), Positives = 47/77 (61%), Gaps = 0/77 (0%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD ++D++ L + FS +GN+ S KV ++ S+G+GFV Y + A A+ ++NG +IG
Sbjct 339 LDDSVDDEKLKEMFSEYGNVTSSKVMLNPQGMSRGFGFVAYSNPEEALRALSEMNGKMIG 398
Query 61 GKTVYVGPFITRSERES 77
K +Y+ + +R +
Sbjct 399 RKPLYIALAQRKEDRRA 415
Score = 48.5 bits (114), Expect = 7e-06, Method: Composition-based stats.
Identities = 38/141 (26%), Positives = 66/141 (46%), Gaps = 8/141 (5%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD + L+D F N++S +V D+ RS GY ++++ + A A+E +N +
Sbjct 56 LDPKVTEAHLFDLFKHVANVVSVRVCRDQNRRSLGYAYINFSNPNDAYRAMEALNYTPLF 115
Query 61 GKTVYVGPFITRSERE-SAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQD 119
+ + I S R+ S ++ N+++KNL D ++L FG I S V D
Sbjct 116 DRPIR----IMLSNRDPSTRLSGKGNIFIKNLDAS-IDNKALFETFSSFGTILSCKVAMD 170
Query 120 PKGRP--FAFCNYAEHSAANA 138
GR + F + + +A A
Sbjct 171 VTGRSKGYGFVQFEKEESAQA 191
> ath:AT1G49760 PAB8; PAB8 (POLY(A) BINDING PROTEIN 8); RNA binding
/ translation initiation factor; K13126 polyadenylate-binding
protein
Length=671
Score = 133 bits (334), Expect = 2e-31, Method: Composition-based stats.
Identities = 67/133 (50%), Positives = 95/133 (71%), Gaps = 4/133 (3%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD++ID+KAL++TFS FG ILSCKV VD + +SKGYGFV Y+++ +A+ AI+K+NGML+
Sbjct 140 LDKSIDHKALHETFSAFGPILSCKVAVDPSGQSKGYGFVQYDTDEAAQGAIDKLNGMLLN 199
Query 61 GKTVYVGPFITRSERESAGVN-RFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQD 119
K VYVGPF+ + +R+ +G +FTNVY+KNL SD E L + FG TS V+ +D
Sbjct 200 DKQVYVGPFVHKLQRDPSGEKVKFTNVYVKNLSESLSD-EELNKVFGEFGVTTSCVIMRD 258
Query 120 PKGRP--FAFCNY 130
+G+ F F N+
Sbjct 259 GEGKSKGFGFVNF 271
Score = 73.6 bits (179), Expect = 2e-13, Method: Composition-based stats.
Identities = 45/135 (33%), Positives = 68/135 (50%), Gaps = 14/135 (10%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
L ++ ++ L F FG SC + D +SKG+GFV++E+ A A++ +NG
Sbjct 231 LSESLSDEELNKVFGEFGVTTSCVIMRDGEGKSKGFGFVNFENSDDAARAVDALNGKTFD 290
Query 61 GKTVYVGPFITRSERESAGVNRF-------------TNVYLKNLPRCWSDPESLRPILEP 107
K +VG +SERE+ +F +N+Y+KNL +D + LR P
Sbjct 291 DKEWFVGKAQKKSERETELKQKFEQSLKEAADKSQGSNLYVKNLDESVTD-DKLREHFAP 349
Query 108 FGEITSLVVKQDPKG 122
FG ITS V +DP G
Sbjct 350 FGTITSCKVMRDPSG 364
Score = 61.6 bits (148), Expect = 7e-10, Method: Composition-based stats.
Identities = 41/141 (29%), Positives = 68/141 (48%), Gaps = 7/141 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVD-EANRSKGYGFVHYESEASARNAIEKVNGMLI 59
LD + + L++ F+ G ++S +V D RS GYG+V+Y + A A+ ++N M +
Sbjct 52 LDATVTDSQLFEAFTQAGQVVSVRVCRDMTTRRSLGYGYVNYATPQDASRALNELNFMAL 111
Query 60 GGKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQD 119
G+ + V + +GV N+++KNL + D ++L FG I S V D
Sbjct 112 NGRAIRVMYSVRDPSLRKSGVG---NIFIKNLDKS-IDHKALHETFSAFGPILSCKVAVD 167
Query 120 PKGRP--FAFCNYAEHSAANA 138
P G+ + F Y AA
Sbjct 168 PSGQSKGYGFVQYDTDEAAQG 188
Score = 55.5 bits (132), Expect = 5e-08, Method: Composition-based stats.
Identities = 30/92 (32%), Positives = 51/92 (55%), Gaps = 0/92 (0%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD ++ + L + F+ FG I SCKV D + S+G GFV + + A AI ++NG +I
Sbjct 334 LDESVTDDKLREHFAPFGTITSCKVMRDPSGVSRGSGFVAFSTPEEATRAITEMNGKMIV 393
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLP 92
K +YV + +R++ +F+ + N+P
Sbjct 394 TKPLYVALAQRKEDRKARLQAQFSQMRPVNMP 425
> xla:495336 hypothetical LOC495336
Length=711
Score = 132 bits (331), Expect = 5e-31, Method: Composition-based stats.
Identities = 72/141 (51%), Positives = 92/141 (65%), Gaps = 6/141 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
L + I+ K LYDTFSLFG ILSCK+ +DE SKGYGFVH+E+E A+ AI+KVN M I
Sbjct 97 LAKTIEQKELYDTFSLFGRILSCKIAMDENGNSKGYGFVHFENEECAKRAIQKVNNMSIC 156
Query 61 GKTVYVGPFITRSERESAG-VNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQD 119
GK VYVG FI RS+R+S +F N+Y+KN P +D E L+ + FGEI S V +D
Sbjct 157 GKVVYVGNFIPRSDRKSQNRKQKFNNIYVKNFPP-ETDDEKLKEMFTEFGEIKSACVMKD 215
Query 120 PKGRP--FAFCNY--AEHSAA 136
+G+ F F Y EH+ A
Sbjct 216 SEGKSKGFGFVCYLNPEHAEA 236
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 42/136 (30%), Positives = 62/136 (45%), Gaps = 15/136 (11%)
Query 6 DNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKTVY 65
D++ L + F+ FG I S V D +SKG+GFV Y + A A+ ++G IGG+++Y
Sbjct 193 DDEKLKEMFTEFGEIKSACVMKDSEGKSKGFGFVCYLNPEHAEAAVAAMHGKEIGGRSLY 252
Query 66 VGPFITRSERESAGVNRF--------------TNVYLKNLPRCWSDPESLRPILEPFGEI 111
+ ER+ R N+Y+KNL D E L+ I +G I
Sbjct 253 ASRAQRKEERQEELKLRLEKQKAERRSKYVSNVNLYVKNLDDEIDD-ERLKEIFSKYGPI 311
Query 112 TSLVVKQDPKGRPFAF 127
+S V D R F
Sbjct 312 SSAKVMTDSNNRSKGF 327
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 29/77 (37%), Positives = 43/77 (55%), Gaps = 0/77 (0%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD ID++ L + FS +G I S KV D NRSKG+GFV + + A A+ + NG +
Sbjct 292 LDDEIDDERLKEIFSKYGPISSAKVMTDSNNRSKGFGFVCFTNPEQATKAVTEANGRVEY 351
Query 61 GKTVYVGPFITRSERES 77
K +YV + +R +
Sbjct 352 SKPLYVAIAQRKEDRRA 368
Score = 42.4 bits (98), Expect = 5e-04, Method: Composition-based stats.
Identities = 38/140 (27%), Positives = 65/140 (46%), Gaps = 9/140 (6%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANR-SKGYGFVHYESEASARNAIEKVNGMLI 59
L +I++ L FS G + V D +R S GYG+V++E A A+E++N ++
Sbjct 9 LHPDINDDQLRMKFSEIGPVAVAHVCRDVTSRKSLGYGYVNFEDPKDAERALEQMNYEVV 68
Query 60 GGKTVYVGPFITRSERE-SAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQ 118
G+ + I S+R+ S + N+++KNL + E L FG I S +
Sbjct 69 MGRPIR----IMWSQRDPSLRKSGLGNIFIKNLAKTIEQKE-LYDTFSLFGRILSCKIAM 123
Query 119 DPKG--RPFAFCNYAEHSAA 136
D G + + F ++ A
Sbjct 124 DENGNSKGYGFVHFENEECA 143
> xla:444284 pabpc4, MGC80927, PABP, ePAB, ePABP; poly(A) binding
protein, cytoplasmic 4 (inducible form); K13126 polyadenylate-binding
protein
Length=626
Score = 131 bits (330), Expect = 5e-31, Method: Composition-based stats.
Identities = 72/142 (50%), Positives = 91/142 (64%), Gaps = 7/142 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD++IDNKALYDTFS FGNILSCKV DE N SKGY FVH+E++ +A AIEK+NGML+
Sbjct 106 LDKSIDNKALYDTFSAFGNILSCKVVCDE-NGSKGYAFVHFETQDAADRAIEKMNGMLLN 164
Query 61 GKTVYVGPFITRSERES---AGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK 117
+ V+VG F R ERE+ A FTNVY+KN D E L+ +G+ S+ V
Sbjct 165 DRKVFVGRFKCRREREAELGAKAKEFTNVYIKNFGEDMDD-ERLKETFSKYGKTLSVKVM 223
Query 118 QDPKGRP--FAFCNYAEHSAAN 137
DP G+ F F ++ H AN
Sbjct 224 TDPSGKSKGFGFVSFERHEDAN 245
Score = 80.1 bits (196), Expect = 2e-15, Method: Composition-based stats.
Identities = 49/147 (33%), Positives = 77/147 (52%), Gaps = 15/147 (10%)
Query 4 NIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKT 63
++D++ L +TFS +G LS KV D + +SKG+GFV +E A A++ +NG + GK
Sbjct 201 DMDDERLKETFSKYGKTLSVKVMTDPSGKSKGFGFVSFERHEDANKAVDDMNGKDVNGKI 260
Query 64 VYVGPFITRSERESAGVNRF-------------TNVYLKNLPRCWSDPESLRPILEPFGE 110
++VG + ER++ RF N+Y+KNL D E LR PFG
Sbjct 261 MFVGRAQKKVERQAELKRRFEQLKQERISRYQGVNLYIKNLDDTIDD-EKLRKEFSPFGS 319
Query 111 ITSL-VVKQDPKGRPFAFCNYAEHSAA 136
ITS V+ ++ + + F F ++ A
Sbjct 320 ITSAKVMLEEGRSKGFGFVCFSSPEEA 346
Score = 62.4 bits (150), Expect = 4e-10, Method: Composition-based stats.
Identities = 35/91 (38%), Positives = 54/91 (59%), Gaps = 5/91 (5%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD ID++ L FS FG+I S KV ++E RSKG+GFV + S A A+ ++NG ++G
Sbjct 301 LDDTIDDEKLRKEFSPFGSITSAKVMLEEG-RSKGFGFVCFSSPEEATKAVTEMNGRIVG 359
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNL 91
K +YV + ER++ TN Y++ +
Sbjct 360 SKPLYVALAQRKEERKA----HLTNQYMQRI 386
Score = 52.8 bits (125), Expect = 4e-07, Method: Composition-based stats.
Identities = 45/143 (31%), Positives = 72/143 (50%), Gaps = 14/143 (9%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDE-ANRSKGYGFVHYESEASARNAIEKVNGMLI 59
L ++ LY+ FS G +LS +V D RS GY +V+++ A A A++ +N +I
Sbjct 18 LHPDVTEAMLYEKFSPAGPVLSIRVCRDMITRRSLGYAYVNFQQPADAERALDTMNFDVI 77
Query 60 GGKTVYVGPFITRSERE----SAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLV 115
GK + I S+R+ +GV NV++KNL + D ++L FG I S
Sbjct 78 KGKPIR----IMWSQRDPSLRKSGVG---NVFIKNLDKS-IDNKALYDTFSAFGNILSCK 129
Query 116 VKQDPKG-RPFAFCNYAEHSAAN 137
V D G + +AF ++ AA+
Sbjct 130 VVCDENGSKGYAFVHFETQDAAD 152
> dre:393856 pabpc1b, MGC77608, pabpc1, zgc:77608; poly A binding
protein, cytoplasmic 1 b; K13126 polyadenylate-binding protein
Length=634
Score = 131 bits (329), Expect = 8e-31, Method: Composition-based stats.
Identities = 72/141 (51%), Positives = 91/141 (64%), Gaps = 7/141 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD++IDNKALYDTFS FGNILSCKV DE N SKGYGFVH+E++ +A AIEK+NGML+
Sbjct 106 LDKSIDNKALYDTFSAFGNILSCKVVCDE-NGSKGYGFVHFETQEAAERAIEKMNGMLLN 164
Query 61 GKTVYVGPFITRSERES---AGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK 117
+ V+VG F +R ERE+ A FTNVY+KN D + L+ I +G S+ V
Sbjct 165 DRKVFVGRFKSRKEREAELGARAKEFTNVYIKNFGEDMDD-DKLKDIFSKYGNAMSIRVM 223
Query 118 QDPKG--RPFAFCNYAEHSAA 136
D G R F F ++ H A
Sbjct 224 TDENGKSRGFGFVSFERHEDA 244
Score = 80.1 bits (196), Expect = 2e-15, Method: Composition-based stats.
Identities = 49/147 (33%), Positives = 76/147 (51%), Gaps = 15/147 (10%)
Query 4 NIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKT 63
++D+ L D FS +GN +S +V DE +S+G+GFV +E A+ A++++NG + GK
Sbjct 201 DMDDDKLKDIFSKYGNAMSIRVMTDENGKSRGFGFVSFERHEDAQRAVDEMNGKEMNGKL 260
Query 64 VYVGPFITRSERESAGVNRF-------------TNVYLKNLPRCWSDPESLRPILEPFGE 110
+YVG + ER++ +F N+Y+KNL D E LR PFG
Sbjct 261 IYVGRAQKKVERQTELKRKFEQMKQDRMTRYQGVNLYVKNLDDGIDD-ERLRKEFSPFGT 319
Query 111 ITSLVVKQD-PKGRPFAFCNYAEHSAA 136
ITS V D + + F F ++ A
Sbjct 320 ITSAKVMMDGGRSKGFGFVCFSSPEEA 346
Score = 62.8 bits (151), Expect = 3e-10, Method: Composition-based stats.
Identities = 37/108 (34%), Positives = 58/108 (53%), Gaps = 8/108 (7%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD ID++ L FS FG I S KV +D RSKG+GFV + S A A+ ++NG ++
Sbjct 301 LDDGIDDERLRKEFSPFGTITSAKVMMD-GGRSKGFGFVCFSSPEEATKAVTEMNGRIVA 359
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPF 108
K +YV + ER++ TN Y++ + + P P++ P+
Sbjct 360 TKPLYVALAQRKEERQA----HLTNQYMQRMASVRAVPN---PVINPY 400
Score = 51.6 bits (122), Expect = 7e-07, Method: Composition-based stats.
Identities = 44/142 (30%), Positives = 71/142 (50%), Gaps = 14/142 (9%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDE-ANRSKGYGFVHYESEASARNAIEKVNGMLI 59
L +++ LY+ FS G ILS +V D RS GY +V+++ A A A++ +N +I
Sbjct 18 LHQDVTEAMLYEKFSPAGAILSIRVCRDMITRRSLGYAYVNFQQPADAERALDTMNFDVI 77
Query 60 GGKTVYVGPFITRSERE----SAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLV 115
G+ V I S+R+ +GV N+++KNL + D ++L FG I S
Sbjct 78 KGRPVR----IMWSQRDPSLRKSGVG---NIFIKNLDKS-IDNKALYDTFSAFGNILSCK 129
Query 116 VKQDPKG-RPFAFCNYAEHSAA 136
V D G + + F ++ AA
Sbjct 130 VVCDENGSKGYGFVHFETQEAA 151
> mmu:230721 Pabpc4, MGC11665, MGC6685; poly(A) binding protein,
cytoplasmic 4; K13126 polyadenylate-binding protein
Length=660
Score = 131 bits (329), Expect = 8e-31, Method: Composition-based stats.
Identities = 72/142 (50%), Positives = 94/142 (66%), Gaps = 7/142 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD++IDNKALYDTFS FGNILSCKV DE N SKGY FVH+E++ +A AIEK+NGML+
Sbjct 106 LDKSIDNKALYDTFSAFGNILSCKVVCDE-NGSKGYAFVHFETQEAADKAIEKMNGMLLN 164
Query 61 GKTVYVGPFITRSERES---AGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK 117
+ V+VG F +R ERE+ A FTNVY+KN D +L+ + FG+ S+ V
Sbjct 165 DRKVFVGRFKSRKEREAELGAKAKEFTNVYIKNFGE-EVDDGNLKELFSQFGKTLSVKVM 223
Query 118 QDPKGRP--FAFCNYAEHSAAN 137
+D G+ F F +Y +H AN
Sbjct 224 RDSSGKSKGFGFVSYEKHEDAN 245
Score = 81.3 bits (199), Expect = 8e-16, Method: Composition-based stats.
Identities = 51/146 (34%), Positives = 75/146 (51%), Gaps = 15/146 (10%)
Query 5 IDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKTV 64
+D+ L + FS FG LS KV D + +SKG+GFV YE A A+E++NG + GK +
Sbjct 202 VDDGNLKELFSQFGKTLSVKVMRDSSGKSKGFGFVSYEKHEDANKAVEEMNGKEMSGKAI 261
Query 65 YVGPFITRSERESAGVNRF-------------TNVYLKNLPRCWSDPESLRPILEPFGEI 111
+VG + ER++ +F N+Y+KNL D E LR PFG I
Sbjct 262 FVGRAQKKVERQAELKRKFEQLKQERISRYQGVNLYIKNLDDTIDD-EKLRREFSPFGSI 320
Query 112 TSL-VVKQDPKGRPFAFCNYAEHSAA 136
TS V+ +D + + F F ++ A
Sbjct 321 TSAKVMLEDGRSKGFGFVCFSSPEEA 346
Score = 60.1 bits (144), Expect = 2e-09, Method: Composition-based stats.
Identities = 36/100 (36%), Positives = 57/100 (57%), Gaps = 5/100 (5%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD ID++ L FS FG+I S KV +++ RSKG+GFV + S A A+ ++NG ++G
Sbjct 301 LDDTIDDEKLRREFSPFGSITSAKVMLEDG-RSKGFGFVCFSSPEEATKAVTEMNGRIVG 359
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPES 100
K +YV + ER++ TN Y++ + + P S
Sbjct 360 SKPLYVALAQRKEERKA----HLTNQYMQRVAGMRALPAS 395
Score = 51.6 bits (122), Expect = 8e-07, Method: Composition-based stats.
Identities = 41/139 (29%), Positives = 68/139 (48%), Gaps = 6/139 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDE-ANRSKGYGFVHYESEASARNAIEKVNGMLI 59
L ++ LY+ FS G +LS +V D RS GY +V+++ A A A++ +N ++
Sbjct 18 LHSDVTEAMLYEKFSPAGPVLSIRVCRDMITRRSLGYAYVNFQQPADAERALDTMNFDVM 77
Query 60 GGKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQD 119
GK + + +GV NV++KNL + D ++L FG I S V D
Sbjct 78 KGKPIRIMWSQRDPSLRKSGVG---NVFIKNLDKS-IDNKALYDTFSAFGNILSCKVVCD 133
Query 120 PKG-RPFAFCNYAEHSAAN 137
G + +AF ++ AA+
Sbjct 134 ENGSKGYAFVHFETQEAAD 152
> dre:327625 pabpc1l, fi20g03, pab-2, wu:fi20g03, zgc:55855; poly(A)
binding protein, cytoplasmic 1-like; K13126 polyadenylate-binding
protein
Length=620
Score = 130 bits (328), Expect = 9e-31, Method: Compositional matrix adjust.
Identities = 73/141 (51%), Positives = 91/141 (64%), Gaps = 7/141 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
+D +IDNKALYDTFS FGNILSCKV DE N SKGYGFVH+E++ +A AIE +NGML+
Sbjct 106 MDESIDNKALYDTFSAFGNILSCKVVCDE-NGSKGYGFVHFETQEAANRAIETMNGMLLN 164
Query 61 GKTVYVGPFITRSERES---AGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK 117
+ V+VG F +R ERE+ A FTNVY+KN D E L+ I FG+ S+ V
Sbjct 165 DRKVFVGHFKSRKEREAEMGAKAVEFTNVYIKNFGEDI-DSEKLKNIFTEFGKTLSVCVM 223
Query 118 QDPKGRP--FAFCNYAEHSAA 136
D +GR F F N+ H A
Sbjct 224 TDERGRSRGFGFVNFVNHGDA 244
Score = 75.5 bits (184), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 49/142 (34%), Positives = 72/142 (50%), Gaps = 15/142 (10%)
Query 4 NIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKT 63
+ID++ L + F+ FG LS V DE RS+G+GFV++ + AR A+ ++NG + G+
Sbjct 201 DIDSEKLKNIFTEFGKTLSVCVMTDERGRSRGFGFVNFVNHGDARRAVTEMNGKELNGRV 260
Query 64 VYVGPFITRSERESAGVNRF-------------TNVYLKNLPRCWSDPESLRPILEPFGE 110
+YVG R ER+ +F N+Y+KNL D E LR P+G
Sbjct 261 LYVGRAQKRLERQGELKRKFEQIKQERIQRYQGVNLYVKNLDDSIDD-EKLRKEFAPYGT 319
Query 111 ITSLVVKQD-PKGRPFAFCNYA 131
ITS V D R F F ++
Sbjct 320 ITSAKVMTDGGHSRGFGFVCFS 341
Score = 54.7 bits (130), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 32/98 (32%), Positives = 52/98 (53%), Gaps = 5/98 (5%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD +ID++ L F+ +G I S KV D S+G+GFV + S A A+ ++NG ++
Sbjct 301 LDDSIDDEKLRKEFAPYGTITSAKVMTD-GGHSRGFGFVCFSSPEEATKAVTEMNGRIVS 359
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDP 98
K +YV + ER++ TN Y++ L + P
Sbjct 360 TKPLYVALAQRKEERKAI----LTNQYIQRLASIRAIP 393
> xla:398221 pabpc1l-a, MGC82273, ePAB, epabp-a, pabpc1l1; poly(A)
binding protein, cytoplasmic 1-like; K13126 polyadenylate-binding
protein
Length=629
Score = 130 bits (326), Expect = 2e-30, Method: Composition-based stats.
Identities = 73/141 (51%), Positives = 88/141 (62%), Gaps = 7/141 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD +IDNKALYDTFS FGNILSCKV DE + S+GYGFVH+E+ +A AI+ +NGML+
Sbjct 106 LDESIDNKALYDTFSAFGNILSCKVVCDE-HGSRGYGFVHFETHEAANRAIQTMNGMLLN 164
Query 61 GKTVYVGPFITRSERE---SAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK 117
+ V+VG F +R ERE A V FTNVY+KN D + LR I FG S+ V
Sbjct 165 DRKVFVGHFKSRRERELEYGAKVMEFTNVYIKNFGEDMDD-KRLREIFSAFGNTLSVKVM 223
Query 118 QDPKGRP--FAFCNYAEHSAA 136
D GR F F NY H A
Sbjct 224 MDDSGRSRGFGFVNYGNHEEA 244
Score = 82.0 bits (201), Expect = 5e-16, Method: Composition-based stats.
Identities = 51/147 (34%), Positives = 81/147 (55%), Gaps = 15/147 (10%)
Query 4 NIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKT 63
++D+K L + FS FGN LS KV +D++ RS+G+GFV+Y + A+ A+ ++NG + G+
Sbjct 201 DMDDKRLREIFSAFGNTLSVKVMMDDSGRSRGFGFVNYGNHEEAQKAVSEMNGKEVNGRM 260
Query 64 VYVGPFITRSERES-----------AGVNRF--TNVYLKNLPRCWSDPESLRPILEPFGE 110
+YVG R ER+S +NR+ N+Y+KNL D + LR P+G
Sbjct 261 IYVGRAQKRIERQSELKRKFEQIKQERINRYQGVNLYVKNLDDGIDD-DRLRKEFLPYGT 319
Query 111 ITSL-VVKQDPKGRPFAFCNYAEHSAA 136
ITS V+ + + F F ++ A
Sbjct 320 ITSAKVMTEGGHSKGFGFVCFSSPEEA 346
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 37/121 (30%), Positives = 59/121 (48%), Gaps = 8/121 (6%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD ID+ L F +G I S KV + E SKG+GFV + S A A+ ++NG ++
Sbjct 301 LDDGIDDDRLRKEFLPYGTITSAKV-MTEGGHSKGFGFVCFSSPEEATKAVTEMNGRIVS 359
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQDP 120
K +YV + ER++ TN Y++ L + P P+L F + + + P
Sbjct 360 TKPLYVALAQRKEERKAI----LTNQYMQRLATMRAMPG---PLLGSFQQPANYFLSAMP 412
Query 121 K 121
+
Sbjct 413 Q 413
> mmu:18458 Pabpc1, PABP, Pabp1, PabpI, Pabpl1, ePAB; poly(A)
binding protein, cytoplasmic 1; K13126 polyadenylate-binding
protein
Length=636
Score = 129 bits (324), Expect = 3e-30, Method: Composition-based stats.
Identities = 72/141 (51%), Positives = 91/141 (64%), Gaps = 7/141 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD++IDNKALYDTFS FGNILSCKV DE N SKGYGFVH+E++ +A AIEK+NGML+
Sbjct 106 LDKSIDNKALYDTFSAFGNILSCKVVCDE-NGSKGYGFVHFETQEAAERAIEKMNGMLLN 164
Query 61 GKTVYVGPFITRSERES---AGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK 117
+ V+VG F +R ERE+ A FTNVY+KN D E L+ + FG S+ V
Sbjct 165 DRKVFVGRFKSRKEREAELGARAKEFTNVYIKNFGEDMDD-ERLKELFGKFGPALSVKVM 223
Query 118 QDPKGRP--FAFCNYAEHSAA 136
D G+ F F ++ H A
Sbjct 224 TDESGKSKGFGFVSFERHEDA 244
Score = 77.8 bits (190), Expect = 9e-15, Method: Composition-based stats.
Identities = 49/147 (33%), Positives = 77/147 (52%), Gaps = 15/147 (10%)
Query 4 NIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKT 63
++D++ L + F FG LS KV DE+ +SKG+GFV +E A+ A++++NG + GK
Sbjct 201 DMDDERLKELFGKFGPALSVKVMTDESGKSKGFGFVSFERHEDAQKAVDEMNGKELNGKQ 260
Query 64 VYVGPFITRSERESAGVNRF-------------TNVYLKNLPRCWSDPESLRPILEPFGE 110
+YVG + ER++ +F N+Y+KNL D E LR PFG
Sbjct 261 IYVGRAQKKVERQTELKRKFEQMKQDRITRYQGVNLYVKNLDDGIDD-ERLRKEFSPFGT 319
Query 111 ITSL-VVKQDPKGRPFAFCNYAEHSAA 136
ITS V+ + + + F F ++ A
Sbjct 320 ITSAKVMMEGGRSKGFGFVCFSSPEEA 346
Score = 62.0 bits (149), Expect = 6e-10, Method: Composition-based stats.
Identities = 37/108 (34%), Positives = 58/108 (53%), Gaps = 8/108 (7%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD ID++ L FS FG I S KV + E RSKG+GFV + S A A+ ++NG ++
Sbjct 301 LDDGIDDERLRKEFSPFGTITSAKVMM-EGGRSKGFGFVCFSSPEEATKAVTEMNGRIVA 359
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPF 108
K +YV + ER++ TN Y++ + + P P++ P+
Sbjct 360 TKPLYVALAQRKEERQA----HLTNQYMQRMASVRAVPN---PVINPY 400
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 42/138 (30%), Positives = 66/138 (47%), Gaps = 6/138 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDE-ANRSKGYGFVHYESEASARNAIEKVNGMLI 59
L ++ LY+ FS G ILS +V D RS GY +V+++ A A A++ +N +I
Sbjct 18 LHPDVTEAMLYEKFSPAGPILSIRVCRDMITRRSLGYAYVNFQQPADAERALDTMNFDVI 77
Query 60 GGKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQD 119
GK V + +GV N+++KNL + D ++L FG I S V D
Sbjct 78 KGKPVRIMWSQRDPSLRKSGVG---NIFIKNLDKS-IDNKALYDTFSAFGNILSCKVVCD 133
Query 120 PKG-RPFAFCNYAEHSAA 136
G + + F ++ AA
Sbjct 134 ENGSKGYGFVHFETQEAA 151
> hsa:26986 PABPC1, PAB1, PABP, PABP1, PABPC2, PABPL1; poly(A)
binding protein, cytoplasmic 1; K13126 polyadenylate-binding
protein
Length=636
Score = 129 bits (324), Expect = 3e-30, Method: Composition-based stats.
Identities = 72/141 (51%), Positives = 91/141 (64%), Gaps = 7/141 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD++IDNKALYDTFS FGNILSCKV DE N SKGYGFVH+E++ +A AIEK+NGML+
Sbjct 106 LDKSIDNKALYDTFSAFGNILSCKVVCDE-NGSKGYGFVHFETQEAAERAIEKMNGMLLN 164
Query 61 GKTVYVGPFITRSERES---AGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK 117
+ V+VG F +R ERE+ A FTNVY+KN D E L+ + FG S+ V
Sbjct 165 DRKVFVGRFKSRKEREAELGARAKEFTNVYIKNFGEDMDD-ERLKDLFGKFGPALSVKVM 223
Query 118 QDPKGRP--FAFCNYAEHSAA 136
D G+ F F ++ H A
Sbjct 224 TDESGKSKGFGFVSFERHEDA 244
Score = 79.7 bits (195), Expect = 3e-15, Method: Composition-based stats.
Identities = 50/147 (34%), Positives = 77/147 (52%), Gaps = 15/147 (10%)
Query 4 NIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKT 63
++D++ L D F FG LS KV DE+ +SKG+GFV +E A+ A++++NG + GK
Sbjct 201 DMDDERLKDLFGKFGPALSVKVMTDESGKSKGFGFVSFERHEDAQKAVDEMNGKELNGKQ 260
Query 64 VYVGPFITRSERESAGVNRF-------------TNVYLKNLPRCWSDPESLRPILEPFGE 110
+YVG + ER++ +F N+Y+KNL D E LR PFG
Sbjct 261 IYVGRAQKKVERQTELKRKFEQMKQDRITRYQGVNLYVKNLDDGIDD-ERLRKEFSPFGT 319
Query 111 ITSL-VVKQDPKGRPFAFCNYAEHSAA 136
ITS V+ + + + F F ++ A
Sbjct 320 ITSAKVMMEGGRSKGFGFVCFSSPEEA 346
Score = 62.0 bits (149), Expect = 6e-10, Method: Composition-based stats.
Identities = 37/108 (34%), Positives = 58/108 (53%), Gaps = 8/108 (7%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD ID++ L FS FG I S KV + E RSKG+GFV + S A A+ ++NG ++
Sbjct 301 LDDGIDDERLRKEFSPFGTITSAKVMM-EGGRSKGFGFVCFSSPEEATKAVTEMNGRIVA 359
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPF 108
K +YV + ER++ TN Y++ + + P P++ P+
Sbjct 360 TKPLYVALAQRKEERQA----HLTNQYMQRMASVRAVPN---PVINPY 400
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 45/142 (31%), Positives = 70/142 (49%), Gaps = 14/142 (9%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDE-ANRSKGYGFVHYESEASARNAIEKVNGMLI 59
L ++ LY+ FS G ILS +V D RS GY +V+++ A A A++ +N +I
Sbjct 18 LHPDVTEAMLYEKFSPAGPILSIRVCRDMITRRSLGYAYVNFQQPADAERALDTMNFDVI 77
Query 60 GGKTVYVGPFITRSERE----SAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLV 115
GK V I S+R+ +GV N+++KNL + D ++L FG I S
Sbjct 78 KGKPVR----IMWSQRDPSLRKSGVG---NIFIKNLDKS-IDNKALYDTFSAFGNILSCK 129
Query 116 VKQDPKG-RPFAFCNYAEHSAA 136
V D G + + F ++ AA
Sbjct 130 VVCDENGSKGYGFVHFETQEAA 151
> dre:606498 pabpc1a, pabpc1, wu:fb16a02, wu:fi19b08, wu:fj12d09,
wu:fj61f06, zgc:109879; poly A binding protein, cytoplasmic
1 a; K13126 polyadenylate-binding protein
Length=634
Score = 128 bits (321), Expect = 6e-30, Method: Composition-based stats.
Identities = 72/141 (51%), Positives = 90/141 (63%), Gaps = 7/141 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD++IDNKALYDTFS FGNILSCKV DE N SKGYGFVH+E+ +A AIEK+NGML+
Sbjct 106 LDKSIDNKALYDTFSAFGNILSCKVVCDE-NGSKGYGFVHFETHEAAERAIEKMNGMLLN 164
Query 61 GKTVYVGPFITRSERES---AGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK 117
+ V+VG F +R ERE+ A FTNVY+KN D E L+ I +G S+ V
Sbjct 165 DRKVFVGRFKSRKEREAEMGARAKEFTNVYIKNFGEDMDD-EKLKEIFCKYGPALSIRVM 223
Query 118 QDPKGRP--FAFCNYAEHSAA 136
D G+ F F ++ H A
Sbjct 224 TDDSGKSKGFGFVSFERHEDA 244
Score = 75.5 bits (184), Expect = 4e-14, Method: Composition-based stats.
Identities = 47/147 (31%), Positives = 77/147 (52%), Gaps = 15/147 (10%)
Query 4 NIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKT 63
++D++ L + F +G LS +V D++ +SKG+GFV +E A+ A++++NG + GK
Sbjct 201 DMDDEKLKEIFCKYGPALSIRVMTDDSGKSKGFGFVSFERHEDAQRAVDEMNGKEMNGKQ 260
Query 64 VYVGPFITRSERESAGVNRF-------------TNVYLKNLPRCWSDPESLRPILEPFGE 110
VYVG + ER++ +F N+Y+KNL D E LR PFG
Sbjct 261 VYVGRAQKKGERQTELKRKFEQMKQDRMTRYQGVNLYVKNLDDGLDD-ERLRKEFSPFGT 319
Query 111 ITSL-VVKQDPKGRPFAFCNYAEHSAA 136
ITS V+ + + + F F ++ A
Sbjct 320 ITSAKVMMEGGRSKGFGFVCFSSPEEA 346
Score = 59.7 bits (143), Expect = 2e-09, Method: Composition-based stats.
Identities = 36/108 (33%), Positives = 58/108 (53%), Gaps = 8/108 (7%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD +D++ L FS FG I S KV + E RSKG+GFV + S A A+ ++NG ++
Sbjct 301 LDDGLDDERLRKEFSPFGTITSAKVMM-EGGRSKGFGFVCFSSPEEATKAVTEMNGRIVA 359
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPF 108
K +YV + ER++ T+ Y++ + + P P+L P+
Sbjct 360 TKPLYVALAQRKEERQA----HLTSQYMQRMASVRAVPN---PVLNPY 400
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 45/142 (31%), Positives = 71/142 (50%), Gaps = 14/142 (9%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVD-EANRSKGYGFVHYESEASARNAIEKVNGMLI 59
L ++ LY+ FS G ILS +V D RS GY +V+++ A A A++ +N +I
Sbjct 18 LHPDVTEAMLYEKFSPAGPILSIRVCRDMMTRRSLGYAYVNFQQPADAERALDTMNFDVI 77
Query 60 GGKTVYVGPFITRSERE----SAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLV 115
G+ V I S+R+ +GV N+++KNL + D ++L FG I S
Sbjct 78 KGRPVR----IMWSQRDPSLRKSGVG---NIFIKNLDKS-IDNKALYDTFSAFGNILSCK 129
Query 116 VKQDPKG-RPFAFCNYAEHSAA 136
V D G + + F ++ H AA
Sbjct 130 VVCDENGSKGYGFVHFETHEAA 151
> mmu:18459 Pabpc2, PABP, PABP+, Pabp2; poly(A) binding protein,
cytoplasmic 2; K13126 polyadenylate-binding protein
Length=628
Score = 127 bits (319), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 71/142 (50%), Positives = 92/142 (64%), Gaps = 7/142 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
L++ IDNKALYDTFS FGNILSCKV DE N SKG+GFVH+E+E +A AIEK+NGML+
Sbjct 106 LNKTIDNKALYDTFSAFGNILSCKVVSDE-NGSKGHGFVHFETEEAAERAIEKMNGMLLN 164
Query 61 GKTVYVGPFITRSERES---AGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK 117
+ V+VG F ++ ERE+ G FTNVY+KN D E+L + FG+I S+ V
Sbjct 165 DRKVFVGRFKSQKEREAELGTGTKEFTNVYIKNFGDRMDD-ETLNGLFGRFGQILSVKVM 223
Query 118 QDPKGRP--FAFCNYAEHSAAN 137
D G+ F F ++ H A
Sbjct 224 TDEGGKSKGFGFVSFERHEDAQ 245
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 47/141 (33%), Positives = 74/141 (52%), Gaps = 15/141 (10%)
Query 5 IDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKTV 64
+D++ L F FG ILS KV DE +SKG+GFV +E A+ A++++NG + GK +
Sbjct 202 MDDETLNGLFGRFGQILSVKVMTDEGGKSKGFGFVSFERHEDAQKAVDEMNGKELNGKHI 261
Query 65 YVGPFITRSERESAGVNRF-------------TNVYLKNLPRCWSDPESLRPILEPFGEI 111
YVG + +R + ++F N+Y+KNL D E L+ PFG I
Sbjct 262 YVGRAQKKDDRHTELKHKFEQVTQDKSIRYQGINLYVKNLDDGIDD-ERLQKEFSPFGTI 320
Query 112 TSL-VVKQDPKGRPFAFCNYA 131
TS V+ + + + F F ++
Sbjct 321 TSTKVMTEGGRSKGFGFVCFS 341
Score = 65.5 bits (158), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 37/103 (35%), Positives = 55/103 (53%), Gaps = 5/103 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD ID++ L FS FG I S KV + E RSKG+GFV + S A A+ ++NG ++
Sbjct 301 LDDGIDDERLQKEFSPFGTITSTKV-MTEGGRSKGFGFVCFSSPEEATKAVSEMNGRIVA 359
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRP 103
K +YV + ER++ TN Y++ + S P + P
Sbjct 360 TKPLYVALAQRKEERQA----HLTNQYIQRMASVRSGPNPVNP 398
Score = 48.9 bits (115), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 47/143 (32%), Positives = 67/143 (46%), Gaps = 16/143 (11%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDE-ANRSKGYGFVHYESEASARNAIEKVNGMLI 59
L ++ LY+ FS G ILS +V D RS GY V++E A A A++ +N +I
Sbjct 18 LHPDVTEAMLYEKFSSAGPILSIRVYRDVITRRSLGYASVNFEQPADAERALDTMNFDVI 77
Query 60 GGKTVYV-----GPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSL 114
GK V + P + RS GV NV++KNL + D ++L FG I S
Sbjct 78 KGKPVRIMWSQRDPSLRRS-----GVG---NVFIKNLNKTI-DNKALYDTFSAFGNILSC 128
Query 115 VVKQDPKG-RPFAFCNYAEHSAA 136
V D G + F ++ AA
Sbjct 129 KVVSDENGSKGHGFVHFETEEAA 151
> xla:443777 pabpc1l-b, MGC81363, epab, epabp-b, pabpc1l1; poly(A)
binding protein, cytoplasmic 1-like; K13126 polyadenylate-binding
protein
Length=629
Score = 126 bits (317), Expect = 2e-29, Method: Composition-based stats.
Identities = 71/141 (50%), Positives = 88/141 (62%), Gaps = 7/141 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD +IDNKALYDTFS FG+ILSCKV DE S+GYGFVH+E++ +A AI+ +NGML+
Sbjct 106 LDDSIDNKALYDTFSAFGDILSCKVVCDEYG-SRGYGFVHFETQEAANRAIQTMNGMLLN 164
Query 61 GKTVYVGPFITRSERE---SAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK 117
+ V+VG F +R ERE A V FTNVY+KN D + L+ I FG S+ V
Sbjct 165 DRKVFVGHFKSRRERELEYGAKVMEFTNVYIKNFGEDMDD-KRLKEIFSAFGNTLSVKVM 223
Query 118 QDPKGRP--FAFCNYAEHSAA 136
D GR F F NY H A
Sbjct 224 MDNSGRSRGFGFVNYGNHEEA 244
Score = 81.3 bits (199), Expect = 1e-15, Method: Composition-based stats.
Identities = 51/147 (34%), Positives = 79/147 (53%), Gaps = 15/147 (10%)
Query 4 NIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKT 63
++D+K L + FS FGN LS KV +D + RS+G+GFV+Y + A+ A+ ++NG + G+
Sbjct 201 DMDDKRLKEIFSAFGNTLSVKVMMDNSGRSRGFGFVNYGNHEEAQKAVTEMNGKEVNGRM 260
Query 64 VYVGPFITRSERE-----------SAGVNRF--TNVYLKNLPRCWSDPESLRPILEPFGE 110
VYVG R ER+ +NR+ N+Y+KNL D + LR P+G
Sbjct 261 VYVGRAQKRIERQGELKRKFEQIKQERINRYQGVNLYVKNLDDGIDD-DRLRKEFSPYGT 319
Query 111 ITSL-VVKQDPKGRPFAFCNYAEHSAA 136
ITS V+ + + F F ++ A
Sbjct 320 ITSTKVMTEGGHSKGFGFVCFSSPEEA 346
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 40/127 (31%), Positives = 62/127 (48%), Gaps = 8/127 (6%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD ID+ L FS +G I S KV + E SKG+GFV + S A A+ ++NG ++
Sbjct 301 LDDGIDDDRLRKEFSPYGTITSTKV-MTEGGHSKGFGFVCFSSPEEATKAVTEMNGRIVS 359
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQDP 120
K +YV + ER++ TN Y++ L + P P+L F + + + P
Sbjct 360 TKPLYVALAQRKEERKAI----LTNQYMQRLATMRAMPG---PLLGSFQQPANYFLPTMP 412
Query 121 KGRPFAF 127
+ AF
Sbjct 413 QPSNRAF 419
> hsa:80336 PABPC1L, C20orf119, FLJ11840, FLJ30809, FLJ42053,
PABPC1L1, dJ1069P2.3, ePAB; poly(A) binding protein, cytoplasmic
1-like; K13126 polyadenylate-binding protein
Length=614
Score = 126 bits (317), Expect = 2e-29, Method: Composition-based stats.
Identities = 68/141 (48%), Positives = 94/141 (66%), Gaps = 7/141 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
L+ +IDNKALYDTFS FGNILSCKV DE + S+G+GFVH+E+ +A+ AI +NGML+
Sbjct 106 LEDSIDNKALYDTFSTFGNILSCKVACDE-HGSRGFGFVHFETHEAAQQAINTMNGMLLN 164
Query 61 GKTVYVGPFITRSERES---AGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK 117
+ V+VG F +R ERE+ A FTN+Y+KNLP D + L+ + FG++ S+ V
Sbjct 165 DRKVFVGHFKSRREREAELGARALEFTNIYVKNLP-VDVDEQGLQDLFSQFGKMLSVKVM 223
Query 118 QDPKG--RPFAFCNYAEHSAA 136
+D G R F F N+ +H A
Sbjct 224 RDNSGHSRCFGFVNFEKHEEA 244
Score = 71.6 bits (174), Expect = 7e-13, Method: Composition-based stats.
Identities = 46/147 (31%), Positives = 72/147 (48%), Gaps = 15/147 (10%)
Query 4 NIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKT 63
++D + L D FS FG +LS KV D + S+ +GFV++E A+ A+ +NG + G+
Sbjct 201 DVDEQGLQDLFSQFGKMLSVKVMRDNSGHSRCFGFVNFEKHEEAQKAVVHMNGKEVSGRL 260
Query 64 VYVGPFITRSERESAGVNRF-------------TNVYLKNLPRCWSDPESLRPILEPFGE 110
+Y G R ER++ RF N+Y+KNL D + LR P+G
Sbjct 261 LYAGRAQKRVERQNELKRRFEQMKQDRLRRYQGVNLYVKNLDDSIDD-DKLRKEFSPYGV 319
Query 111 ITSL-VVKQDPKGRPFAFCNYAEHSAA 136
ITS V+ + + F F ++ A
Sbjct 320 ITSAKVMTEGGHSKGFGFVCFSSPEEA 346
Score = 58.5 bits (140), Expect = 7e-09, Method: Composition-based stats.
Identities = 37/100 (37%), Positives = 55/100 (55%), Gaps = 7/100 (7%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD +ID+ L FS +G I S KV + E SKG+GFV + S A A+ ++NG ++G
Sbjct 301 LDDSIDDDKLRKEFSPYGVITSAKV-MTEGGHSKGFGFVCFSSPEEATKAVTEMNGRIVG 359
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLP--RCWSDP 98
K +YV + ER++ TN Y++ L R S+P
Sbjct 360 TKPLYVALAQRKEERKAI----LTNQYMQRLSTMRTLSNP 395
Score = 55.5 bits (132), Expect = 6e-08, Method: Composition-based stats.
Identities = 45/142 (31%), Positives = 71/142 (50%), Gaps = 14/142 (9%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEAN-RSKGYGFVHYESEASARNAIEKVNGMLI 59
L ++ LY+ FS G ILS +V D A RS GY +++++ A A A++ +N ++
Sbjct 18 LHPDVTEAMLYEKFSPAGPILSIRVCRDVATRRSLGYAYINFQQPADAERALDTMNFEML 77
Query 60 GGKTVYVGPFITRSERE----SAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLV 115
G+ + I S+R+ +GV N+++KNL D ++L FG I S
Sbjct 78 KGQPIR----IMWSQRDPGLRKSGVG---NIFIKNLEDS-IDNKALYDTFSTFGNILSCK 129
Query 116 VKQDPKG-RPFAFCNYAEHSAA 136
V D G R F F ++ H AA
Sbjct 130 VACDEHGSRGFGFVHFETHEAA 151
> xla:379896 pabpc1-a, MGC53109, pab1, pabp, pabp1, pabpc1, pabpc2;
poly(A) binding protein, cytoplasmic 1; K13126 polyadenylate-binding
protein
Length=633
Score = 126 bits (317), Expect = 2e-29, Method: Composition-based stats.
Identities = 70/141 (49%), Positives = 92/141 (65%), Gaps = 7/141 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD++IDNKALYDTFS FGNILSCKV DE N SKGYGFVH+E++ +A AI+K+NGML+
Sbjct 106 LDKSIDNKALYDTFSAFGNILSCKVVCDE-NGSKGYGFVHFETQEAAERAIDKMNGMLLN 164
Query 61 GKTVYVGPFITRSERES---AGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK 117
+ V+VG F +R ERE+ A FTNVY+KN +D E L+ + +G S+ V
Sbjct 165 DRKVFVGRFKSRKEREAELGARAKEFTNVYIKNFGDDMND-ERLKEMFGKYGPALSVKVM 223
Query 118 QDPKGRP--FAFCNYAEHSAA 136
D G+ F F ++ H A
Sbjct 224 TDDNGKSKGFGFVSFERHEDA 244
Score = 66.6 bits (161), Expect = 2e-11, Method: Composition-based stats.
Identities = 44/147 (29%), Positives = 76/147 (51%), Gaps = 15/147 (10%)
Query 4 NIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKT 63
+++++ L + F +G LS KV D+ +SKG+GFV +E A+ A++++ G + GK+
Sbjct 201 DMNDERLKEMFGKYGPALSVKVMTDDNGKSKGFGFVSFERHEDAQKAVDEMYGKDMNGKS 260
Query 64 VYVGPFITRSERESAGVNRF-------------TNVYLKNLPRCWSDPESLRPILEPFGE 110
++VG + ER++ +F N+Y+KNL D E LR PFG
Sbjct 261 MFVGRAQKKVERQTELKRKFEQMNQDRITRYQGVNLYVKNLDDGIDD-ERLRKEFLPFGT 319
Query 111 ITSL-VVKQDPKGRPFAFCNYAEHSAA 136
ITS V+ + + + F F ++ A
Sbjct 320 ITSAKVMMEGGRSKGFGFVCFSSPEEA 346
Score = 56.2 bits (134), Expect = 3e-08, Method: Composition-based stats.
Identities = 35/108 (32%), Positives = 56/108 (51%), Gaps = 9/108 (8%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD ID++ L F FG I S KV + E RSKG+GFV + S A A+ ++NG ++
Sbjct 301 LDDGIDDERLRKEFLPFGTITSAKVMM-EGGRSKGFGFVCFSSPEEATKAVTEMNGRIVA 359
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPF 108
K +YV + ER++ TN Y++ + + P++ P+
Sbjct 360 TKPLYVALAQRKEERQA----HLTNQYMQRM----ASVRVPNPVINPY 399
Score = 51.2 bits (121), Expect = 1e-06, Method: Composition-based stats.
Identities = 44/142 (30%), Positives = 71/142 (50%), Gaps = 14/142 (9%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDE-ANRSKGYGFVHYESEASARNAIEKVNGMLI 59
L +++ LY+ FS G ILS +V D RS GY +V+++ A A A++ +N +I
Sbjct 18 LHQDVTEAMLYEKFSPAGPILSIRVCRDMITRRSLGYAYVNFQQPADAERALDTMNFDVI 77
Query 60 GGKTVYVGPFITRSERE----SAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLV 115
G+ V I S+R+ +GV N+++KNL + D ++L FG I S
Sbjct 78 KGRPVR----IMWSQRDPSLRKSGVG---NIFIKNLDKS-IDNKALYDTFSAFGNILSCK 129
Query 116 VKQDPKG-RPFAFCNYAEHSAA 136
V D G + + F ++ AA
Sbjct 130 VVCDENGSKGYGFVHFETQEAA 151
> xla:432140 pabpc1-b, MGC130736, MGC79060, pab1, pabp, pabp1,
pabpc2; poly(A) binding protein, cytoplasmic 1; K13126 polyadenylate-binding
protein
Length=633
Score = 126 bits (316), Expect = 2e-29, Method: Composition-based stats.
Identities = 71/141 (50%), Positives = 90/141 (63%), Gaps = 7/141 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD++IDNKALYDTFS FGNILSCKV DE N SKGYGFVH+E++ +A AI+K+NGML+
Sbjct 106 LDKSIDNKALYDTFSAFGNILSCKVVCDE-NGSKGYGFVHFETQEAAERAIDKMNGMLLN 164
Query 61 GKTVYVGPFITRSERES---AGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK 117
+ V+VG F +R ERE+ A FTNVY+KN D E L+ +G S+ V
Sbjct 165 DRKVFVGRFKSRKEREAELGARAKEFTNVYIKNFGEDMDD-ERLKEWFGQYGAALSVKVM 223
Query 118 QDPKG--RPFAFCNYAEHSAA 136
D G R F F ++ H A
Sbjct 224 TDDHGKSRGFGFVSFERHEDA 244
Score = 71.2 bits (173), Expect = 9e-13, Method: Composition-based stats.
Identities = 45/147 (30%), Positives = 75/147 (51%), Gaps = 15/147 (10%)
Query 4 NIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKT 63
++D++ L + F +G LS KV D+ +S+G+GFV +E A+ A++ +NG + GK
Sbjct 201 DMDDERLKEWFGQYGAALSVKVMTDDHGKSRGFGFVSFERHEDAQKAVDDMNGKDLNGKA 260
Query 64 VYVGPFITRSERESAGVNRF-------------TNVYLKNLPRCWSDPESLRPILEPFGE 110
++VG + ER++ +F N+Y+KNL D E LR PFG
Sbjct 261 IFVGRAQKKVERQTELKRKFEQMKQDRITRYQGVNLYVKNLDDGIDD-ERLRKEFTPFGS 319
Query 111 ITSL-VVKQDPKGRPFAFCNYAEHSAA 136
ITS V+ + + + F F ++ A
Sbjct 320 ITSAKVMMEGGRSKGFGFVCFSSPEEA 346
Score = 58.2 bits (139), Expect = 7e-09, Method: Composition-based stats.
Identities = 35/108 (32%), Positives = 58/108 (53%), Gaps = 9/108 (8%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD ID++ L F+ FG+I S KV + E RSKG+GFV + S A A+ ++NG ++
Sbjct 301 LDDGIDDERLRKEFTPFGSITSAKVMM-EGGRSKGFGFVCFSSPEEATKAVTEMNGRIVA 359
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPF 108
K +YV + ER++ TN Y++ + + P++ P+
Sbjct 360 TKPLYVALAQRKEERQA----HLTNQYMQRM----ASVRVPNPVINPY 399
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 45/142 (31%), Positives = 70/142 (49%), Gaps = 14/142 (9%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDE-ANRSKGYGFVHYESEASARNAIEKVNGMLI 59
L ++ LY+ FS G ILS +V D RS GY +V+++ A A A++ +N +I
Sbjct 18 LHPDVTEAMLYEKFSPAGPILSIRVCRDMITRRSLGYAYVNFQQPADAERALDTMNFDVI 77
Query 60 GGKTVYVGPFITRSERE----SAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLV 115
GK V I S+R+ +GV N+++KNL + D ++L FG I S
Sbjct 78 KGKPVR----IMWSQRDPSLRKSGVG---NIFIKNLDKS-IDNKALYDTFSAFGNILSCK 129
Query 116 VKQDPKG-RPFAFCNYAEHSAA 136
V D G + + F ++ AA
Sbjct 130 VVCDENGSKGYGFVHFETQEAA 151
> mmu:100503296 polyadenylate-binding protein 4-like
Length=234
Score = 124 bits (311), Expect = 8e-29, Method: Compositional matrix adjust.
Identities = 67/130 (51%), Positives = 87/130 (66%), Gaps = 5/130 (3%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD++IDNKALYDTFS FGNILSCKV DE N SKGY FVH+E++ +A AIEK+NGML+
Sbjct 106 LDKSIDNKALYDTFSAFGNILSCKVVCDE-NGSKGYAFVHFETQEAADKAIEKMNGMLLN 164
Query 61 GKTVYVGPFITRSERES---AGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK 117
+ V+VG F +R ERE+ A FTNVY+KN D +L+ + FG+ S+ V
Sbjct 165 DRKVFVGRFKSRKEREAELGAKAKEFTNVYIKNFGEEVDDG-NLKELFSQFGKTLSVKVM 223
Query 118 QDPKGRPFAF 127
+D G+ F
Sbjct 224 RDSSGKSKGF 233
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 44/143 (30%), Positives = 72/143 (50%), Gaps = 14/143 (9%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDE-ANRSKGYGFVHYESEASARNAIEKVNGMLI 59
L ++ LY+ FS G +LS +V D RS GY +V+++ A A A++ +N ++
Sbjct 18 LHSDVTEAMLYEKFSPAGPVLSIRVCRDMITRRSLGYAYVNFQQPADAERALDTMNFDVM 77
Query 60 GGKTVYVGPFITRSERE----SAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLV 115
GK + I S+R+ +GV NV++KNL + D ++L FG I S
Sbjct 78 KGKPIR----IMWSQRDPSLRKSGVG---NVFIKNLDKSI-DNKALYDTFSAFGNILSCK 129
Query 116 VKQDPKG-RPFAFCNYAEHSAAN 137
V D G + +AF ++ AA+
Sbjct 130 VVCDENGSKGYAFVHFETQEAAD 152
Score = 32.7 bits (73), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 5 IDNKALYDTFSLFGNILSCKVGVDEANRSKGYG 37
+D+ L + FS FG LS KV D + +SKG+G
Sbjct 202 VDDGNLKELFSQFGKTLSVKVMRDSSGKSKGFG 234
> sce:YER165W PAB1; Pab1p; K13126 polyadenylate-binding protein
Length=577
Score = 123 bits (309), Expect = 2e-28, Method: Composition-based stats.
Identities = 63/141 (44%), Positives = 92/141 (65%), Gaps = 6/141 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
L +IDNKALYDTFS+FG+ILS K+ DE +SKG+GFVH+E E +A+ AI+ +NGML+
Sbjct 133 LHPDIDNKALYDTFSVFGDILSSKIATDENGKSKGFGFVHFEEEGAAKEAIDALNGMLLN 192
Query 61 GKTVYVGPFITRSERESA---GVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK 117
G+ +YV P ++R ER+S +TN+Y+KN+ +D E + + FG I S ++
Sbjct 193 GQEIYVAPHLSRKERDSQLEETKAHYTNLYVKNINSETTD-EQFQELFAKFGPIVSASLE 251
Query 118 QDPKG--RPFAFCNYAEHSAA 136
+D G + F F NY +H A
Sbjct 252 KDADGKLKGFGFVNYEKHEDA 272
Score = 56.2 bits (134), Expect = 3e-08, Method: Composition-based stats.
Identities = 39/134 (29%), Positives = 62/134 (46%), Gaps = 14/134 (10%)
Query 7 NKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKTVYV 66
++ + F+ FG I+S + D + KG+GFV+YE A A+E +N + G+ +YV
Sbjct 232 DEQFQELFAKFGPIVSASLEKDADGKLKGFGFVNYEKHEDAVKAVEALNDSELNGEKLYV 291
Query 67 GPFITRSER-------------ESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITS 113
G ++ER E + N+++KNL D E L P+G ITS
Sbjct 292 GRAQKKNERMHVLKKQYEAYRLEKMAKYQGVNLFVKNLDDSVDD-EKLEEEFAPYGTITS 350
Query 114 LVVKQDPKGRPFAF 127
V + G+ F
Sbjct 351 AKVMRTENGKSKGF 364
Score = 51.2 bits (121), Expect = 1e-06, Method: Composition-based stats.
Identities = 25/67 (37%), Positives = 40/67 (59%), Gaps = 0/67 (0%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD ++D++ L + F+ +G I S KV E +SKG+GFV + + A AI + N ++
Sbjct 329 LDDSVDDEKLEEEFAPYGTITSAKVMRTENGKSKGFGFVCFSTPEEATKAITEKNQQIVA 388
Query 61 GKTVYVG 67
GK +YV
Sbjct 389 GKPLYVA 395
Score = 51.2 bits (121), Expect = 1e-06, Method: Composition-based stats.
Identities = 42/141 (29%), Positives = 71/141 (50%), Gaps = 11/141 (7%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANR-SKGYGFVHYESEASARNAIEKVNGMLI 59
L+ ++ LYD FS G++ S +V D + S GY +V++ + R AIE++N I
Sbjct 45 LEPSVSEAHLYDIFSPIGSVSSIRVCRDAITKTSLGYAYVNFNDHEAGRKAIEQLNYTPI 104
Query 60 GGKTVYVGPFITRSERESAGVNRFT-NVYLKNL-PRCWSDPESLRPILEPFGEITSLVVK 117
G+ I S+R+ + + + N+++KNL P D ++L FG+I S +
Sbjct 105 KGRLCR----IMWSQRDPSLRKKGSGNIFIKNLHPDI--DNKALYDTFSVFGDILSSKIA 158
Query 118 QDPKGRP--FAFCNYAEHSAA 136
D G+ F F ++ E AA
Sbjct 159 TDENGKSKGFGFVHFEEEGAA 179
> ath:AT2G23350 PAB4; PAB4 (POLY(A) BINDING PROTEIN 4); RNA binding
/ translation initiation factor
Length=662
Score = 123 bits (309), Expect = 2e-28, Method: Composition-based stats.
Identities = 63/133 (47%), Positives = 90/133 (67%), Gaps = 4/133 (3%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD+++DNK L++ FS G I+SCKV D +S+GYGFV +++E SA+NAIEK+NG ++
Sbjct 141 LDKSVDNKTLHEAFSGCGTIVSCKVATDHMGQSRGYGFVQFDTEDSAKNAIEKLNGKVLN 200
Query 61 GKTVYVGPFITRSERESAGVN-RFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQD 119
K ++VGPF+ + ERESA +FTNVY+KNL +D E L+ +G I+S VV +D
Sbjct 201 DKQIFVGPFLRKEERESAADKMKFTNVYVKNLSEATTDDE-LKTTFGQYGSISSAVVMRD 259
Query 120 PKG--RPFAFCNY 130
G R F F N+
Sbjct 260 GDGKSRCFGFVNF 272
Score = 68.9 bits (167), Expect = 5e-12, Method: Composition-based stats.
Identities = 48/126 (38%), Positives = 65/126 (51%), Gaps = 14/126 (11%)
Query 10 LYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKTVYVGPF 69
L TF +G+I S V D +S+ +GFV++E+ A A+E +NG K YVG
Sbjct 241 LKTTFGQYGSISSAVVMRDGDGKSRCFGFVNFENPEDAARAVEALNGKKFDDKEWYVGKA 300
Query 70 ITRSERE-----------SAGVNRFT--NVYLKNLPRCWSDPESLRPILEPFGEITSLVV 116
+SERE S G N+F N+Y+KNL +D E LR + FG ITS V
Sbjct 301 QKKSERELELSRRYEQGSSDGGNKFDGLNLYVKNLDDTVTD-EKLRELFAEFGTITSCKV 359
Query 117 KQDPKG 122
+DP G
Sbjct 360 MRDPSG 365
Score = 58.9 bits (141), Expect = 4e-09, Method: Composition-based stats.
Identities = 30/92 (32%), Positives = 51/92 (55%), Gaps = 0/92 (0%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD + ++ L + F+ FG I SCKV D + SKG GFV + + + A + ++NG ++G
Sbjct 335 LDDTVTDEKLRELFAEFGTITSCKVMRDPSGTSKGSGFVAFSAASEASRVLNEMNGKMVG 394
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLP 92
GK +YV + ER + +F+ + +P
Sbjct 395 GKPLYVALAQRKEERRAKLQAQFSQMRPAFIP 426
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 46/142 (32%), Positives = 70/142 (49%), Gaps = 8/142 (5%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEA-NRSKGYGFVHYESEASARNAIEKVNGMLI 59
LD N+ + LYD F+ ++S +V D A N S GYG+V+Y + A A++K+N +
Sbjct 53 LDFNVTDSQLYDYFTEVCQVVSVRVCRDAATNTSLGYGYVNYSNTDDAEKAMQKLNYSYL 112
Query 60 GGKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQD 119
GK + + S +GV N+++KNL + D ++L G I S V D
Sbjct 113 NGKMIRITYSSRDSSARRSGVG---NLFVKNLDKS-VDNKTLHEAFSGCGTIVSCKVATD 168
Query 120 PKG--RPFAFCNY-AEHSAANA 138
G R + F + E SA NA
Sbjct 169 HMGQSRGYGFVQFDTEDSAKNA 190
> mmu:67543 Pabpc6, 4932702K14Rik, AI428050, MGC132900, Pabpc3;
poly(A) binding protein, cytoplasmic 6; K13126 polyadenylate-binding
protein
Length=643
Score = 122 bits (305), Expect = 4e-28, Method: Composition-based stats.
Identities = 69/141 (48%), Positives = 88/141 (62%), Gaps = 7/141 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LDR+ID+K LYDTFS FGNILSCKV DE N SKGYGFVH+E++ A AIEK+NGM +
Sbjct 106 LDRSIDSKTLYDTFSAFGNILSCKVVCDE-NGSKGYGFVHFETQEEAERAIEKMNGMFLN 164
Query 61 GKTVYVGPFITRSERES---AGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK 117
V+VG F +R +R++ A FTNVY+KNL D E L+ + FG S+ V
Sbjct 165 DHKVFVGRFKSRRDRQAELGARAKEFTNVYIKNLGEDMDD-ERLQDLFGRFGPALSVKVM 223
Query 118 QDPKGRP--FAFCNYAEHSAA 136
D G+ F F ++ H A
Sbjct 224 TDESGKSKGFGFVSFERHEDA 244
Score = 80.5 bits (197), Expect = 1e-15, Method: Composition-based stats.
Identities = 53/160 (33%), Positives = 78/160 (48%), Gaps = 25/160 (15%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
L ++D++ L D F FG LS KV DE+ +SKG+GFV +E AR A+E++NG +
Sbjct 198 LGEDMDDERLQDLFGRFGPALSVKVMTDESGKSKGFGFVSFERHEDARKAVEEMNGKDLN 257
Query 61 GKTVYVGPFITRSERESAGVNRF-----------------------TNVYLKNLPRCWSD 97
GK +YVG + ER++ ++F N+Y+KNL D
Sbjct 258 GKQIYVGRAQKKVERQTELKHKFGQMKQDKHKIERVPQDRSVRCKGVNLYVKNLDDGIDD 317
Query 98 PESLRPILEPFGEITSL-VVKQDPKGRPFAFCNYAEHSAA 136
E LR PFG ITS V + + + F F ++ A
Sbjct 318 -ERLRKEFSPFGTITSAKVTMEGGRSKGFGFVCFSSPEEA 356
Score = 63.2 bits (152), Expect = 3e-10, Method: Composition-based stats.
Identities = 37/108 (34%), Positives = 58/108 (53%), Gaps = 8/108 (7%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD ID++ L FS FG I S KV + E RSKG+GFV + S A A+ ++NG ++
Sbjct 311 LDDGIDDERLRKEFSPFGTITSAKVTM-EGGRSKGFGFVCFSSPEEATKAVTEMNGKIVA 369
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPF 108
K +YV + ER++ +N Y++ + + P P++ PF
Sbjct 370 TKPLYVALAQRKEERQA----HLSNQYMQRMASTSAGPN---PVVSPF 410
Score = 46.2 bits (108), Expect = 3e-05, Method: Composition-based stats.
Identities = 44/142 (30%), Positives = 67/142 (47%), Gaps = 14/142 (9%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDE-ANRSKGYGFVHYESEASARNAIEKVNGMLI 59
L ++ LY+ FS G ILS +V D RS GY V+++ A A++ +N +I
Sbjct 18 LHPDVTEAMLYEKFSPAGPILSIRVYRDRITRRSLGYASVNFQQLEDAERALDTMNFDVI 77
Query 60 GGKTVYVGPFITRSERE----SAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLV 115
GK V I S+R+ +GV N+++KNL R D ++L FG I S
Sbjct 78 KGKPVR----IMWSQRDPSLRKSGVG---NIFVKNLDRS-IDSKTLYDTFSAFGNILSCK 129
Query 116 VKQDPKG-RPFAFCNYAEHSAA 136
V D G + + F ++ A
Sbjct 130 VVCDENGSKGYGFVHFETQEEA 151
> hsa:5042 PABPC3, PABP3, PABPL3, tPABP; poly(A) binding protein,
cytoplasmic 3; K13126 polyadenylate-binding protein
Length=631
Score = 120 bits (302), Expect = 1e-27, Method: Composition-based stats.
Identities = 68/141 (48%), Positives = 88/141 (62%), Gaps = 7/141 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD++I+NKALYDT S FGNILSC V DE N SKGYGFVH+E+ +A AI+K+NGML+
Sbjct 106 LDKSINNKALYDTVSAFGNILSCNVVCDE-NGSKGYGFVHFETHEAAERAIKKMNGMLLN 164
Query 61 GKTVYVGPFITRSERES---AGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK 117
G+ V+VG F +R ERE+ A F NVY+KN D E L+ + FG S+ V
Sbjct 165 GRKVFVGQFKSRKEREAELGARAKEFPNVYIKNFGEDMDD-ERLKDLFGKFGPALSVKVM 223
Query 118 QDPKGRP--FAFCNYAEHSAA 136
D G+ F F ++ H A
Sbjct 224 TDESGKSKGFGFVSFERHEDA 244
Score = 79.0 bits (193), Expect = 4e-15, Method: Composition-based stats.
Identities = 50/147 (34%), Positives = 76/147 (51%), Gaps = 15/147 (10%)
Query 4 NIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKT 63
++D++ L D F FG LS KV DE+ +SKG+GFV +E A+ A++++NG + GK
Sbjct 201 DMDDERLKDLFGKFGPALSVKVMTDESGKSKGFGFVSFERHEDAQKAVDEMNGKELNGKQ 260
Query 64 VYVGPFITRSERESAGVNRF-------------TNVYLKNLPRCWSDPESLRPILEPFGE 110
+YVG + ER++ F N+Y+KNL D E LR PFG
Sbjct 261 IYVGRAQKKVERQTELKRTFEQMKQDRITRYQVVNLYVKNLDDGIDD-ERLRKAFSPFGT 319
Query 111 ITSL-VVKQDPKGRPFAFCNYAEHSAA 136
ITS V+ + + + F F ++ A
Sbjct 320 ITSAKVMMEGGRSKGFGFVCFSSPEEA 346
Score = 59.3 bits (142), Expect = 3e-09, Method: Composition-based stats.
Identities = 36/103 (34%), Positives = 54/103 (52%), Gaps = 5/103 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD ID++ L FS FG I S KV + E RSKG+GFV + S A A+ ++NG ++
Sbjct 301 LDDGIDDERLRKAFSPFGTITSAKVMM-EGGRSKGFGFVCFSSPEEATKAVTEMNGRIVA 359
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRP 103
K +YV + ER++ TN Y++ + + P P
Sbjct 360 TKPLYVALAQRKEERQA----YLTNEYMQRMASVRAVPNQRAP 398
> mmu:381404 Pabpc1l, 1810053B01Rik, Epab; poly(A) binding protein,
cytoplasmic 1-like; K13126 polyadenylate-binding protein
Length=607
Score = 113 bits (282), Expect = 2e-25, Method: Composition-based stats.
Identities = 63/141 (44%), Positives = 91/141 (64%), Gaps = 7/141 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
L+ +IDNKALYDTFS FG+ILS KV +E + S+G+GFVH+E+ +A+ AI +NGML+
Sbjct 106 LENSIDNKALYDTFSTFGSILSSKVVYNE-HGSRGFGFVHFETHEAAQKAINTMNGMLLN 164
Query 61 GKTVYVGPFITRSERESAGVNR---FTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK 117
+ V+VG F +R +RE+ R FTN+Y+KNL D + L+ + FG + S+ V
Sbjct 165 DRKVFVGHFKSRQKREAELGARALGFTNIYVKNL-HANVDEQRLQDLFSQFGNMQSVKVM 223
Query 118 QDPKG--RPFAFCNYAEHSAA 136
+D G R F F N+ +H A
Sbjct 224 RDSNGQSRGFGFVNFEKHEEA 244
Score = 81.3 bits (199), Expect = 8e-16, Method: Composition-based stats.
Identities = 50/150 (33%), Positives = 78/150 (52%), Gaps = 15/150 (10%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
L N+D + L D FS FGN+ S KV D +S+G+GFV++E A+ A++ +NG +
Sbjct 198 LHANVDEQRLQDLFSQFGNMQSVKVMRDSNGQSRGFGFVNFEKHEEAQKAVDHMNGKEVS 257
Query 61 GKTVYVGPFITRSERESAGVNRF-------------TNVYLKNLPRCWSDPESLRPILEP 107
G+ +YVG R+ER+S RF N+Y+KNL +D E L+ +
Sbjct 258 GQLLYVGRAQKRAERQSELKRRFEQMKQERQNRYQGVNLYVKNLDDSIND-ERLKEVFST 316
Query 108 FGEITSL-VVKQDPKGRPFAFCNYAEHSAA 136
+G ITS V+ + + F F ++ A
Sbjct 317 YGVITSAKVMTESSHSKGFGFVCFSSPEEA 346
Score = 58.5 bits (140), Expect = 6e-09, Method: Composition-based stats.
Identities = 37/121 (30%), Positives = 65/121 (53%), Gaps = 14/121 (11%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD +I+++ L + FS +G I S KV + E++ SKG+GFV + S A A+ ++NG ++G
Sbjct 301 LDDSINDERLKEVFSTYGVITSAKV-MTESSHSKGFGFVCFSSPEEATKAVTEMNGRIVG 359
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQDP 120
K +YV + ER++ N++ + P P+L F + TS ++ P
Sbjct 360 TKPLYVALAQRKEERKAILTNQY-----RRRPS--------HPVLSSFQQPTSYLLPAVP 406
Query 121 K 121
+
Sbjct 407 Q 407
Score = 58.5 bits (140), Expect = 6e-09, Method: Composition-based stats.
Identities = 45/140 (32%), Positives = 72/140 (51%), Gaps = 10/140 (7%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEAN-RSKGYGFVHYESEASARNAIEKVNGMLI 59
L ++ LY+ FS GNILS +V D A RS GY +++++ A A A++ +N +I
Sbjct 18 LHPDVTESMLYEMFSPIGNILSIRVCRDVATRRSLGYAYINFQQPADAERALDTMNFEVI 77
Query 60 GGKTVYVGPFITRSERESAGVNR--FTNVYLKNLPRCWSDPESLRPILEPFGEI-TSLVV 116
G+ + I S R+ G+ + N+++KNL D ++L FG I +S VV
Sbjct 78 KGQPIR----IMWSHRD-PGLRKSGMGNIFIKNLENS-IDNKALYDTFSTFGSILSSKVV 131
Query 117 KQDPKGRPFAFCNYAEHSAA 136
+ R F F ++ H AA
Sbjct 132 YNEHGSRGFGFVHFETHEAA 151
> hsa:132430 PABPC4L, DKFZp686J06116; poly(A) binding protein,
cytoplasmic 4-like; K13126 polyadenylate-binding protein
Length=428
Score = 108 bits (269), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 60/141 (42%), Positives = 87/141 (61%), Gaps = 7/141 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD++IDNK LY+ FS FG ILS KV D+ SKGY FVH++++++A AIE++NG L+
Sbjct 163 LDKSIDNKTLYEHFSAFGKILSSKVMSDDQG-SKGYAFVHFQNQSAADRAIEEMNGKLLK 221
Query 61 GKTVYVGPFITRSERES---AGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK 117
G V+VG F R +RE+ + + FTNVY+KN D E L+ + +G+ S+ V
Sbjct 222 GCKVFVGRFKNRKDREAELRSKASEFTNVYIKNFGGDMDD-ERLKDVFSKYGKTLSVKVM 280
Query 118 QDPKGRP--FAFCNYAEHSAA 136
D G+ F F ++ H AA
Sbjct 281 TDSSGKSKGFGFVSFDSHEAA 301
Score = 75.5 bits (184), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 43/129 (33%), Positives = 69/129 (53%), Gaps = 14/129 (10%)
Query 4 NIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKT 63
++D++ L D FS +G LS KV D + +SKG+GFV ++S +A+ A+E++NG I G+
Sbjct 258 DMDDERLKDVFSKYGKTLSVKVMTDSSGKSKGFGFVSFDSHEAAKKAVEEMNGRDINGQL 317
Query 64 VYVGPFITRSERESAGVNRF-------------TNVYLKNLPRCWSDPESLRPILEPFGE 110
++VG + ER++ F +Y+KNL D E LR FG
Sbjct 318 IFVGRAQKKVERQAELKQMFEQLKRERIRGCQGVKLYIKNLDDTIDD-EKLRNEFSSFGS 376
Query 111 ITSLVVKQD 119
I+ + V Q+
Sbjct 377 ISRVKVMQE 385
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 42/143 (29%), Positives = 77/143 (53%), Gaps = 14/143 (9%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANR-SKGYGFVHYESEASARNAIEKVNGMLI 59
L ++ L+ FS G +LS ++ D+ R S GY +V++ A A+ A++ +N +I
Sbjct 75 LHADVTEDLLFRKFSTVGPVLSIRICRDQVTRRSLGYAYVNFLQLADAQKALDTMNFDII 134
Query 60 GGKTVYVGPFITRSERES----AGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLV 115
GK++ + S+R++ +G+ NV++KNL + D ++L FG+I S
Sbjct 135 KGKSIR----LMWSQRDAYLRRSGIG---NVFIKNLDKSI-DNKTLYEHFSAFGKILSSK 186
Query 116 VKQDPKG-RPFAFCNYAEHSAAN 137
V D +G + +AF ++ SAA+
Sbjct 187 VMSDDQGSKGYAFVHFQNQSAAD 209
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 40/66 (60%), Gaps = 1/66 (1%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD ID++ L + FS FG+I KV + E +SKG+G + + S A A+ ++NG ++G
Sbjct 358 LDDTIDDEKLRNEFSSFGSISRVKV-MQEEGQSKGFGLICFSSPEDATKAMTEMNGRILG 416
Query 61 GKTVYV 66
K + +
Sbjct 417 SKPLSI 422
> ath:AT1G34140 PAB1; PAB1 (POLY(A) BINDING PROTEIN 1); RNA binding
/ translation initiation factor
Length=407
Score = 107 bits (268), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 63/138 (45%), Positives = 85/138 (61%), Gaps = 5/138 (3%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD +IDNK L D FS FG +LSCKV D + SKGYGFV + S+ S A NG LI
Sbjct 38 LDESIDNKQLCDMFSAFGKVLSCKVARDASGVSKGYGFVQFYSDLSVYTACNFHNGTLIR 97
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQDP 120
+ ++V PF++R + + + V FTNVY+KNL +D + L+ + FGEITS VV +D
Sbjct 98 NQHIHVCPFVSRGQWDKSRV--FTNVYVKNLVETATDAD-LKRLFGEFGEITSAVVMKDG 154
Query 121 KG--RPFAFCNYAEHSAA 136
+G R F F N+ + AA
Sbjct 155 EGKSRRFGFVNFEKAEAA 172
Score = 62.0 bits (149), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 30/66 (45%), Positives = 40/66 (60%), Gaps = 0/66 (0%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD ++DN L + FS FG I SCKV V SKG GFV + + A A+ K+NG ++G
Sbjct 230 LDDSVDNTKLEELFSEFGTITSCKVMVHSNGISKGVGFVEFSTSEEASKAMLKMNGKMVG 289
Query 61 GKTVYV 66
K +YV
Sbjct 290 NKPIYV 295
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 38/121 (31%), Positives = 56/121 (46%), Gaps = 15/121 (12%)
Query 10 LYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKTVYVGPF 69
L F FG I S V D +S+ +GFV++E +A AIEK+NG+++ K ++VG
Sbjct 135 LKRLFGEFGEITSAVVMKDGEGKSRRFGFVNFEKAEAAVTAIEKMNGVVVDEKELHVGRA 194
Query 70 ITRSERESAGVNRF--------------TNVYLKNLPRCWSDPESLRPILEPFGEITSLV 115
++ R +F N+Y+KNL D L + FG ITS
Sbjct 195 QRKTNRTEDLKAKFELEKIIRDMKTRKGMNLYVKNLDDS-VDNTKLEELFSEFGTITSCK 253
Query 116 V 116
V
Sbjct 254 V 254
> mmu:241989 Pabpc4l, C330050A14Rik, EG241989; poly(A) binding
protein, cytoplasmic 4-like; K13126 polyadenylate-binding protein
Length=370
Score = 107 bits (266), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 60/141 (42%), Positives = 85/141 (60%), Gaps = 7/141 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD++IDNK LY+ FS FG I+S KV D SKGYGFVHY+ +A AIE++NG L+
Sbjct 105 LDKSIDNKTLYEHFSPFGTIMSSKVMTD-GEGSKGYGFVHYQDRRAADRAIEEMNGKLLR 163
Query 61 GKTVYVGPFITRSERESAGVNR---FTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK 117
T++V F +R +RE+ ++ FTNVY+KN D E LR + +G+ S+ V
Sbjct 164 ESTLFVARFKSRKDREAELRDKPTEFTNVYIKNFGDDV-DDEKLREVFSKYGQTLSVKVM 222
Query 118 QDPKGRP--FAFCNYAEHSAA 136
+D G+ F F ++ H AA
Sbjct 223 KDATGKSKGFGFVSFDSHEAA 243
Score = 77.0 bits (188), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 44/130 (33%), Positives = 69/130 (53%), Gaps = 14/130 (10%)
Query 4 NIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKT 63
++D++ L + FS +G LS KV D +SKG+GFV ++S +A+NA+E +NG I G+T
Sbjct 200 DVDDEKLREVFSKYGQTLSVKVMKDATGKSKGFGFVSFDSHEAAKNAVEDMNGQDINGQT 259
Query 64 VYVGPFITRSERESAGVNRF-------------TNVYLKNLPRCWSDPESLRPILEPFGE 110
++VG + ER++ F +Y+KNL D E+LR FG
Sbjct 260 IFVGRAQKKVERQAELKEMFEQMKKERIRARQAAKLYIKNLDDTI-DDETLRKEFSVFGS 318
Query 111 ITSLVVKQDP 120
I + V Q+
Sbjct 319 ICRVKVMQEA 328
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 42/143 (29%), Positives = 74/143 (51%), Gaps = 14/143 (9%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDE-ANRSKGYGFVHYESEASARNAIEKVNGMLI 59
L ++ L+ FS G +LS ++ D + RS GY +V++ A+ A+ +N +I
Sbjct 17 LHEDVTEDMLFRKFSTVGPVLSIRICRDLISQRSLGYAYVNFLQVNDAQKALVTMNFDVI 76
Query 60 GGKTVYVGPFITRSERES----AGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLV 115
GK++ + S+R++ +GV NV++KNL + D ++L PFG I S
Sbjct 77 KGKSIR----LMWSQRDACLRRSGVG---NVFIKNLDKSI-DNKTLYEHFSPFGTIMSSK 128
Query 116 VKQDPKG-RPFAFCNYAEHSAAN 137
V D +G + + F +Y + AA+
Sbjct 129 VMTDGEGSKGYGFVHYQDRRAAD 151
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 40/62 (64%), Gaps = 1/62 (1%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD ID++ L FS+FG+I KV + EA +SKG+G + + S +A A+ ++NG ++G
Sbjct 300 LDDTIDDETLRKEFSVFGSICRVKV-MQEAGQSKGFGLICFFSPEAAAKAMAEMNGRILG 358
Query 61 GK 62
K
Sbjct 359 SK 360
> hsa:140886 PABPC5, FLJ51677, PABP5; poly(A) binding protein,
cytoplasmic 5; K13126 polyadenylate-binding protein
Length=382
Score = 97.4 bits (241), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 61/143 (42%), Positives = 84/143 (58%), Gaps = 8/143 (5%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD++IDN+AL+ FS FGNILSCKV D+ N SKGY +VH++S A+A AI +NG+ +
Sbjct 113 LDKSIDNRALFYLFSAFGNILSCKVVCDD-NGSKGYAYVHFDSLAAANRAIWHMNGVRLN 171
Query 61 GKTVYVGPFITRSERESAGVNR----FTNVYLKNLPRCWSDPESLRPILEPFGEITSLVV 116
+ VYVG F ER + R FTNV++KN+ D E L+ + +G S+ V
Sbjct 172 NRQVYVGRFKFPEERAAEVRTRDRATFTNVFVKNIGDDIDD-EKLKELFCEYGPTESVKV 230
Query 117 KQDPKGRP--FAFCNYAEHSAAN 137
+D G+ F F Y H AA
Sbjct 231 IRDASGKSKGFGFVRYETHEAAQ 253
Score = 58.2 bits (139), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 46/150 (30%), Positives = 76/150 (50%), Gaps = 18/150 (12%)
Query 4 NIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKT 63
+ID++ L + F +G S KV D + +SKG+GFV YE+ +A+ A+ ++G I GK
Sbjct 209 DIDDEKLKELFCEYGPTESVKVIRDASGKSKGFGFVRYETHEAAQKAVLDLHGKSIDGKV 268
Query 64 VYVGPFITRSERESAGVNRFTN-------------VYLKNLPRCWSDPESLRPILEPFGE 110
+YVG + ER + RF +Y+KNL +D E L+ FG
Sbjct 269 LYVGRAQKKIERLAELRRRFERLRLKEKSRPPGVPIYIKNLDETIND-EKLKEEFSSFGS 327
Query 111 IT-SLVVKQDPKGRPFA---FCNYAEHSAA 136
I+ + V+ + +G+ F F ++ E + A
Sbjct 328 ISRAKVMMEVGQGKGFGVVCFSSFEEATKA 357
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 41/66 (62%), Gaps = 1/66 (1%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD I+++ L + FS FG+I KV + E + KG+G V + S A A++++NG ++G
Sbjct 309 LDETINDEKLKEEFSSFGSISRAKVMM-EVGQGKGFGVVCFSSFEEATKAVDEMNGRIVG 367
Query 61 GKTVYV 66
K ++V
Sbjct 368 SKPLHV 373
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 40/139 (28%), Positives = 64/139 (46%), Gaps = 6/139 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSK-GYGFVHYESEASARNAIEKVNGMLI 59
LD ++ LY F G + ++ D RS GYG+V++ A A A+ +N LI
Sbjct 25 LDPDVTEDMLYKKFRPAGPLRFTRICRDPVTRSPLGYGYVNFRFPADAEWALNTMNFDLI 84
Query 60 GGKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQD 119
GK + +GV N+++KNL + D +L + FG I S V D
Sbjct 85 NGKPFRLMWSQPDDRLRKSGVG---NIFIKNLDKSI-DNRALFYLFSAFGNILSCKVVCD 140
Query 120 PKG-RPFAFCNYAEHSAAN 137
G + +A+ ++ +AAN
Sbjct 141 DNGSKGYAYVHFDSLAAAN 159
> mmu:93728 Pabpc5, C820015E17, MGC130340, MGC130341; poly(A)
binding protein, cytoplasmic 5; K13126 polyadenylate-binding
protein
Length=381
Score = 97.4 bits (241), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 61/143 (42%), Positives = 81/143 (56%), Gaps = 8/143 (5%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD+ IDN+AL+ FS FGNILSCKV D+ N SKGY +VH++S A+A AI +NG+ +
Sbjct 112 LDKTIDNRALFYLFSAFGNILSCKVVCDD-NGSKGYAYVHFDSLAAANRAIWHMNGVRLN 170
Query 61 GKTVYVGPFITRSERESAGVNR----FTNVYLKNLPRCWSDPESLRPILEPFGEITSLVV 116
+ VYVG F ER + R FTNV++KN D E L + +G S+ V
Sbjct 171 NRQVYVGRFKFPEERAAEVRTRERATFTNVFVKNFGDDIDD-EKLNKLFSEYGPTESVKV 229
Query 117 KQDPKGRP--FAFCNYAEHSAAN 137
+D G+ F F Y H AA
Sbjct 230 IRDATGKSKGFGFVRYETHEAAQ 252
Score = 59.7 bits (143), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 44/147 (29%), Positives = 72/147 (48%), Gaps = 15/147 (10%)
Query 4 NIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKT 63
+ID++ L FS +G S KV D +SKG+GFV YE+ +A+ A+ +++G I GK
Sbjct 208 DIDDEKLNKLFSEYGPTESVKVIRDATGKSKGFGFVRYETHEAAQKAVLELHGKSIDGKV 267
Query 64 VYVGPFITRSERESAGVNRFTN-------------VYLKNLPRCWSDPESLRPILEPFGE 110
+ VG + ER + RF +Y+KNL +D E L+ FG
Sbjct 268 LCVGRAQKKIERLAELRRRFERLKLKEKNRPSGVPIYIKNLDETIND-EKLKEEFSSFGS 326
Query 111 IT-SLVVKQDPKGRPFAFCNYAEHSAA 136
I+ + V+ + +G+ F ++ A
Sbjct 327 ISRAKVMMEVGQGKGFGVVCFSSFEEA 353
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 42/66 (63%), Gaps = 1/66 (1%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD I+++ L + FS FG+I KV + E + KG+G V + S A A++++NG +IG
Sbjct 308 LDETINDEKLKEEFSSFGSISRAKVMM-EVGQGKGFGVVCFSSFEEACKAVDEMNGRIIG 366
Query 61 GKTVYV 66
KT++V
Sbjct 367 SKTLHV 372
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 40/139 (28%), Positives = 64/139 (46%), Gaps = 6/139 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSK-GYGFVHYESEASARNAIEKVNGMLI 59
LD ++ LY F G + ++ D RS GYG+V++ A A A+ +N LI
Sbjct 24 LDPDVTEDMLYKKFRPAGPLRFTRICRDPVTRSPLGYGYVNFRFPADAEWALNTMNFDLI 83
Query 60 GGKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQD 119
GK + +GV N+++KNL + D +L + FG I S V D
Sbjct 84 NGKPFRLMWSQPDDRLRKSGVG---NIFIKNLDKT-IDNRALFYLFSAFGNILSCKVVCD 139
Query 120 PKG-RPFAFCNYAEHSAAN 137
G + +A+ ++ +AAN
Sbjct 140 DNGSKGYAYVHFDSLAAAN 158
> ath:AT2G36660 PAB7; PAB7 (POLY(A) BINDING PROTEIN 7); RNA binding
/ translation initiation factor
Length=609
Score = 96.7 bits (239), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 51/132 (38%), Positives = 78/132 (59%), Gaps = 4/132 (3%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
L ++ N L D F FGNI+SCKV E +S+GYGFV +E E +A AI+ +N ++
Sbjct 119 LPESVTNAVLQDMFKKFGNIVSCKVATLEDGKSRGYGFVQFEQEDAAHAAIQTLNSTIVA 178
Query 61 GKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQDP 120
K +YVG F+ +++R ++TN+Y+KNL S+ + LR FG+I SL + +D
Sbjct 179 DKEIYVGKFMKKTDRVKPE-EKYTNLYMKNLDADVSE-DLLREKFAEFGKIVSLAIAKDE 236
Query 121 KG--RPFAFCNY 130
R +AF N+
Sbjct 237 NRLCRGYAFVNF 248
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 45/140 (32%), Positives = 68/140 (48%), Gaps = 14/140 (10%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD ++ L + F+ FG I+S + DE +GY FV++++ AR A E VNG G
Sbjct 208 LDADVSEDLLREKFAEFGKIVSLAIAKDENRLCRGYAFVNFDNPEDARRAAETVNGTKFG 267
Query 61 GKTVYVGPFITRSERESAGVNRF-------------TNVYLKNLPRCWSDPESLRPILEP 107
K +YVG ++ERE +F +N+Y+KN+ ++ E LR
Sbjct 268 SKCLYVGRAQKKAEREQLLREQFKEKHEEQKMIAKVSNIYVKNVNVAVTE-EELRKHFSQ 326
Query 108 FGEITSLVVKQDPKGRPFAF 127
G ITS + D KG+ F
Sbjct 327 CGTITSTKLMCDEKGKSKGF 346
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 43/133 (32%), Positives = 69/133 (51%), Gaps = 7/133 (5%)
Query 10 LYDTFSLFGNILSCKVGVDEAN-RSKGYGFVHYESEASARNAIEKVNGMLIGGKTVYVGP 68
LYD F+ F ++ S ++ D ++ RS YG+ ++ S A AIEK N L+ GK + V
Sbjct 40 LYDAFAEFKSLTSVRLCKDASSGRSLCYGYANFLSRQDANLAIEKKNNSLLNGKMIRVMW 99
Query 69 FITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVK--QDPKGRPFA 126
+ + GV NV++KNLP ++ L+ + + FG I S V +D K R +
Sbjct 100 SVRAPDARRNGVG---NVFVKNLPESVTNA-VLQDMFKKFGNIVSCKVATLEDGKSRGYG 155
Query 127 FCNYAEHSAANAC 139
F + + AA+A
Sbjct 156 FVQFEQEDAAHAA 168
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 42/81 (51%), Gaps = 0/81 (0%)
Query 5 IDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIGGKTV 64
+ + L FS G I S K+ DE +SKG+GFV + + A +A++ +G + GK +
Sbjct 315 VTEEELRKHFSQCGTITSTKLMCDEKGKSKGFGFVCFSTPEEAIDAVKTFHGQMFHGKPL 374
Query 65 YVGPFITRSERESAGVNRFTN 85
YV + +R+ +F N
Sbjct 375 YVAIAQKKEDRKMQLQVQFGN 395
> ath:AT3G16380 PAB6; PAB6 (POLY(A) BINDING PROTEIN 6); RNA binding
/ translation initiation factor
Length=537
Score = 91.7 bits (226), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 54/133 (40%), Positives = 82/133 (61%), Gaps = 5/133 (3%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD +I + L F FG+ILSCKV V+E +SKG+GFV +++E SA +A ++G ++
Sbjct 119 LDSSITSSCLERMFCPFGSILSCKV-VEENGQSKGFGFVQFDTEQSAVSARSALHGSMVY 177
Query 61 GKTVYVGPFITRSERES-AGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQD 119
GK ++V FI + ER + AG TNVY+KNL +D + L + +G ++S+VV +D
Sbjct 178 GKKLFVAKFINKDERAAMAGNQDSTNVYVKNLIETVTD-DCLHTLFSQYGTVSSVVVMRD 236
Query 120 PKGRP--FAFCNY 130
GR F F N+
Sbjct 237 GMGRSRGFGFVNF 249
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 40/139 (28%), Positives = 68/139 (48%), Gaps = 13/139 (9%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
L + + L+ FS +G + S V D RS+G+GFV++ + +A+ A+E + G+ +G
Sbjct 209 LIETVTDDCLHTLFSQYGTVSSVVVMRDGMGRSRGFGFVNFCNPENAKKAMESLCGLQLG 268
Query 61 GKTVYVGPFITRSERESAGVNRF------------TNVYLKNLPRCWSDPESLRPILEPF 108
K ++VG + + ER +F +N+Y+KNL ++ LR I +
Sbjct 269 SKKLFVGKALKKDERREMLKQKFSDNFIAKPNMRWSNLYVKNLSESMNETR-LREIFGCY 327
Query 109 GEITSLVVKQDPKGRPFAF 127
G+I S V GR F
Sbjct 328 GQIVSAKVMCHENGRSKGF 346
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 34/110 (30%), Positives = 55/110 (50%), Gaps = 11/110 (10%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
L +++ L + F +G I+S KV E RSKG+GFV + + ++ A +NG L+
Sbjct 311 LSESMNETRLREIFGCYGQIVSAKVMCHENGRSKGFGFVCFSNCEESKQAKRYLNGFLVD 370
Query 61 GKTVYVGPFITR-SERESAGVNRFTNVYLKNLPRCW----SDPESLRPIL 105
GK P + R +ER+ + R Y + PR + S P +P+L
Sbjct 371 GK-----PIVVRVAERKEDRIKRLQQ-YFQAQPRQYTQAPSAPSPAQPVL 414
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 45/147 (30%), Positives = 74/147 (50%), Gaps = 16/147 (10%)
Query 1 LDRNIDNKALYDTFSLFGNILS---CKVGVDEANRSKGYGFVHYESEASARNAIEKVNGM 57
L ++ K L D FSL ++S C+ V +S Y +++++S SA NA+ ++N
Sbjct 28 LSPDVTEKDLIDKFSLNVPVVSVHLCRNSV--TGKSMCYAYINFDSPFSASNAMTRLNHS 85
Query 58 LIGGKTVYVGPFITRSERESAGVNR----FTNVYLKNLPRCWSDPESLRPILEPFGEITS 113
+ GK + I S+R+ A R F N+Y+KNL + L + PFG I S
Sbjct 86 DLKGKAMR----IMWSQRDLAYRRRTRTGFANLYVKNLDSSITS-SCLERMFCPFGSILS 140
Query 114 L-VVKQDPKGRPFAFCNY-AEHSAANA 138
VV+++ + + F F + E SA +A
Sbjct 141 CKVVEENGQSKGFGFVQFDTEQSAVSA 167
> mmu:668382 Pabpc1l2b-ps, EG668382, Pabpc1l2b, Rbm32b; poly(A)
binding protein, cytoplasmic 1-like 2B, pseudogene; K13126
polyadenylate-binding protein
Length=229
Score = 86.3 bits (212), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 41/77 (53%), Positives = 54/77 (70%), Gaps = 1/77 (1%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
L + IDNKALY+ FS FGNILSCKV DE KGYGFVH++ + SA AI+ +NGM +
Sbjct 126 LGKTIDNKALYNIFSAFGNILSCKVACDEKG-PKGYGFVHFQKQESAERAIDALNGMFLN 184
Query 61 GKTVYVGPFITRSERES 77
+ ++VG F + ERE+
Sbjct 185 YRKIFVGRFKSHKEREA 201
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 43/138 (31%), Positives = 69/138 (50%), Gaps = 6/138 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANR-SKGYGFVHYESEASARNAIEKVNGMLI 59
L + LY+ FS G ILS ++ D+ R S GY +V+Y+ A+ A+E +N +I
Sbjct 38 LHPEVTESMLYEKFSPAGPILSIRICRDKVTRRSLGYAYVNYQQPVDAKRALETMNFDVI 97
Query 60 GGKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQD 119
G+ V + +GV NV++KNL + D ++L I FG I S V D
Sbjct 98 NGRPVRIMWSQRDPSLRKSGVG---NVFIKNLGKTI-DNKALYNIFSAFGNILSCKVACD 153
Query 120 PKG-RPFAFCNYAEHSAA 136
KG + + F ++ + +A
Sbjct 154 EKGPKGYGFVHFQKQESA 171
> hsa:340529 PABPC1L2A, MGC168104, PABPC1L2B, RBM32A; poly(A)
binding protein, cytoplasmic 1-like 2A; K13126 polyadenylate-binding
protein
Length=200
Score = 86.3 bits (212), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 46/105 (43%), Positives = 65/105 (61%), Gaps = 4/105 (3%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
L + IDNKALY+ FS FGNILSCKV DE KGYGFVH++ + SA AI+ +NGM +
Sbjct 97 LGKTIDNKALYNIFSAFGNILSCKVACDEKG-PKGYGFVHFQKQESAERAIDVMNGMFLN 155
Query 61 GKTVYVGPFITRSERES---AGVNRFTNVYLKNLPRCWSDPESLR 102
+ ++VG F + ERE+ A + T+ +K+ + +LR
Sbjct 156 YRKIFVGRFKSHKEREAERGAWARQSTSADVKDFEEDTDEEATLR 200
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 42/129 (32%), Positives = 67/129 (51%), Gaps = 6/129 (4%)
Query 10 LYDTFSLFGNILSCKVGVDE-ANRSKGYGFVHYESEASARNAIEKVNGMLIGGKTVYVGP 68
LY+ FS G ILS ++ D+ RS GY +V+Y+ A+ A+E +N +I G+ V +
Sbjct 18 LYEKFSPAGPILSIRICRDKITRRSLGYAYVNYQQPVDAKRALETLNFDVIKGRPVRIMW 77
Query 69 FITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQDPKG-RPFAF 127
+GV NV++KNL + D ++L I FG I S V D KG + + F
Sbjct 78 SQRDPSLRKSGVG---NVFIKNLGKT-IDNKALYNIFSAFGNILSCKVACDEKGPKGYGF 133
Query 128 CNYAEHSAA 136
++ + +A
Sbjct 134 VHFQKQESA 142
> hsa:645974 PABPC1L2B, MGC168104, PABPC1L2A, RBM32B; poly(A)
binding protein, cytoplasmic 1-like 2B; K13126 polyadenylate-binding
protein
Length=200
Score = 86.3 bits (212), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 46/105 (43%), Positives = 65/105 (61%), Gaps = 4/105 (3%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
L + IDNKALY+ FS FGNILSCKV DE KGYGFVH++ + SA AI+ +NGM +
Sbjct 97 LGKTIDNKALYNIFSAFGNILSCKVACDEKG-PKGYGFVHFQKQESAERAIDVMNGMFLN 155
Query 61 GKTVYVGPFITRSERES---AGVNRFTNVYLKNLPRCWSDPESLR 102
+ ++VG F + ERE+ A + T+ +K+ + +LR
Sbjct 156 YRKIFVGRFKSHKEREAERGAWARQSTSADVKDFEEDTDEEATLR 200
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 42/129 (32%), Positives = 67/129 (51%), Gaps = 6/129 (4%)
Query 10 LYDTFSLFGNILSCKVGVDE-ANRSKGYGFVHYESEASARNAIEKVNGMLIGGKTVYVGP 68
LY+ FS G ILS ++ D+ RS GY +V+Y+ A+ A+E +N +I G+ V +
Sbjct 18 LYEKFSPAGPILSIRICRDKITRRSLGYAYVNYQQPVDAKRALETLNFDVIKGRPVRIMW 77
Query 69 FITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQDPKG-RPFAF 127
+GV NV++KNL + D ++L I FG I S V D KG + + F
Sbjct 78 SQRDPSLRKSGVG---NVFIKNLGKT-IDNKALYNIFSAFGNILSCKVACDEKGPKGYGF 133
Query 128 CNYAEHSAA 136
++ + +A
Sbjct 134 VHFQKQESA 142
> mmu:67959 Puf60, 2410104I19Rik, 2810454F19Rik, SIAHBP1; poly-U
binding splicing factor 60; K12838 poly(U)-binding-splicing
factor PUF60
Length=499
Score = 60.1 bits (144), Expect = 2e-09, Method: Composition-based stats.
Identities = 38/136 (27%), Positives = 71/136 (52%), Gaps = 17/136 (12%)
Query 14 FSLFGNILSCKVGVDEAN-RSKGYGFVHYESEASARNAIEKVNGMLIGGKTVYVG----- 67
F+ FG I S + D + KG+ FV YE +A+ A+E++N +++GG+ + VG
Sbjct 89 FAPFGPIKSIDMSWDSVTMKHKGFAFVEYEVPEAAQLALEQMNSVMLGGRNIKVGRPSNI 148
Query 68 ----PFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQDP--- 120
P I + E+ NR +Y+ ++ + SD + ++ + E FG+I S + +DP
Sbjct 149 GQAQPIIDQLAEEARAFNR---IYVASVHQDLSD-DDIKSVFEAFGKIKSCTLARDPTTG 204
Query 121 KGRPFAFCNYAEHSAA 136
K + + F Y + ++
Sbjct 205 KHKGYGFIEYEKAQSS 220
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 22/70 (31%), Positives = 39/70 (55%), Gaps = 1/70 (1%)
Query 3 RNIDNKALYDTFSLFGNILSCKVGVDEAN-RSKGYGFVHYESEASARNAIEKVNGMLIGG 61
+++ + + F FG I SC + D + KGYGF+ YE S+++A+ +N +GG
Sbjct 175 QDLSDDDIKSVFEAFGKIKSCTLARDPTTGKHKGYGFIEYEKAQSSQDAVSSMNLFDLGG 234
Query 62 KTVYVGPFIT 71
+ + VG +T
Sbjct 235 QYLRVGKAVT 244
> hsa:22827 PUF60, FIR, FLJ31379, RoBPI, SIAHBP1; poly-U binding
splicing factor 60KDa; K12838 poly(U)-binding-splicing factor
PUF60
Length=516
Score = 60.1 bits (144), Expect = 2e-09, Method: Composition-based stats.
Identities = 38/136 (27%), Positives = 71/136 (52%), Gaps = 17/136 (12%)
Query 14 FSLFGNILSCKVGVDEAN-RSKGYGFVHYESEASARNAIEKVNGMLIGGKTVYVG----- 67
F+ FG I S + D + KG+ FV YE +A+ A+E++N +++GG+ + VG
Sbjct 106 FAPFGPIKSIDMSWDSVTMKHKGFAFVEYEVPEAAQLALEQMNSVMLGGRNIKVGRPSNI 165
Query 68 ----PFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQDP--- 120
P I + E+ NR +Y+ ++ + SD + ++ + E FG+I S + +DP
Sbjct 166 GQAQPIIDQLAEEARAFNR---IYVASVHQDLSD-DDIKSVFEAFGKIKSCTLARDPTTG 221
Query 121 KGRPFAFCNYAEHSAA 136
K + + F Y + ++
Sbjct 222 KHKGYGFIEYEKAQSS 237
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 22/70 (31%), Positives = 39/70 (55%), Gaps = 1/70 (1%)
Query 3 RNIDNKALYDTFSLFGNILSCKVGVDEAN-RSKGYGFVHYESEASARNAIEKVNGMLIGG 61
+++ + + F FG I SC + D + KGYGF+ YE S+++A+ +N +GG
Sbjct 192 QDLSDDDIKSVFEAFGKIKSCTLARDPTTGKHKGYGFIEYEKAQSSQDAVSSMNLFDLGG 251
Query 62 KTVYVGPFIT 71
+ + VG +T
Sbjct 252 QYLRVGKAVT 261
> tgo:TGME49_024580 splicing factor, putative (EC:3.1.2.15); K12831
splicing factor 3B subunit 4
Length=576
Score = 59.3 bits (142), Expect = 4e-09, Method: Composition-based stats.
Identities = 32/87 (36%), Positives = 50/87 (57%), Gaps = 4/87 (4%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVD-EANRSKGYGFVHYESEASARNAIEKVNGMLI 59
LD ++D K +YDTFS FGNI+S K+ D E S+G+GFV +++ ++ A+ +NG I
Sbjct 123 LDPDVDEKTIYDTFSAFGNIISAKIMRDPETGLSRGFGFVSFDTFEASDAALAAMNGQFI 182
Query 60 GGKTVYVGPFI---TRSERESAGVNRF 83
+ ++V TR ER + R
Sbjct 183 CNRPIHVSYAYKKDTRGERHGSAAERL 209
Score = 48.1 bits (113), Expect = 9e-06, Method: Composition-based stats.
Identities = 38/143 (26%), Positives = 70/143 (48%), Gaps = 11/143 (7%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDE-ANRSKGYGFVHYESEASARNAIEKVNGMLI 59
LD +D+ L++ F G + + V D+ +GYGFV + +E A A++ +N + +
Sbjct 36 LDSQVDDDLLWELFVQCGPVRTVSVPRDKLTGNHQGYGFVEFRNEVDADYALKLMNMVKL 95
Query 60 GGKTVYVGPFITRSERESAGVNRFTNVYLKNL-PRCWSDPESLRPILEPFGEITSLVVKQ 118
GK + + +S ++ + NV+L NL P D +++ FG I S + +
Sbjct 96 YGKALR----LNKSAQDRRNFDVGANVFLGNLDPDV--DEKTIYDTFSAFGNIISAKIMR 149
Query 119 DPK---GRPFAFCNYAEHSAANA 138
DP+ R F F ++ A++A
Sbjct 150 DPETGLSRGFGFVSFDTFEASDA 172
> dre:562370 puf60a, fc21a05, wu:fc21a05, wu:fi43a08; poly-U binding
splicing factor a; K12838 poly(U)-binding-splicing factor
PUF60
Length=518
Score = 58.5 bits (140), Expect = 7e-09, Method: Composition-based stats.
Identities = 39/136 (28%), Positives = 69/136 (50%), Gaps = 17/136 (12%)
Query 14 FSLFGNILSCKVGVDEAN-RSKGYGFVHYESEASARNAIEKVNGMLIGGKTVYVG----- 67
F+ FG I S + D + KG+ FV YE +A+ A+E++N +++GG+ + VG
Sbjct 112 FAPFGPIKSIDMSWDSVTLKHKGFAFVEYEVPEAAQLALEQMNSVMLGGRNIKVGRPSNI 171
Query 68 ----PFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQDP--- 120
P I + E+ NR +Y+ ++ SD + ++ + E FG I S + +DP
Sbjct 172 GQAQPIIDQLAEEARAFNR---IYVASVHPDLSD-DDIKSVFEAFGRIKSCSLARDPTTG 227
Query 121 KGRPFAFCNYAEHSAA 136
K + + F Y + +A
Sbjct 228 KHKGYGFIEYDKAQSA 243
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 22/69 (31%), Positives = 38/69 (55%), Gaps = 1/69 (1%)
Query 4 NIDNKALYDTFSLFGNILSCKVGVDEAN-RSKGYGFVHYESEASARNAIEKVNGMLIGGK 62
++ + + F FG I SC + D + KGYGF+ Y+ SA++A+ +N +GG+
Sbjct 199 DLSDDDIKSVFEAFGRIKSCSLARDPTTGKHKGYGFIEYDKAQSAQDAVSSMNLFDLGGQ 258
Query 63 TVYVGPFIT 71
+ VG +T
Sbjct 259 YLRVGKAVT 267
> pfa:PF14_0194 spliceosome-associated protein, putative; K12831
splicing factor 3B subunit 4
Length=484
Score = 58.2 bits (139), Expect = 7e-09, Method: Composition-based stats.
Identities = 32/86 (37%), Positives = 48/86 (55%), Gaps = 3/86 (3%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEANRSKGYGFVHYESEASARNAIEKVNGMLIG 60
LD +D K L+D FS FG I++ KV +E + SKG+GF+ Y++ S+ AIE +N I
Sbjct 114 LDDEVDEKMLFDIFSSFGQIMTVKVMRNEDDTSKGHGFISYDNFESSDLAIENMNNQFIC 173
Query 61 GKTVYVGPFI---TRSERESAGVNRF 83
K V++ ++ ER RF
Sbjct 174 NKKVHISYAFKKDSKGERHGTAAERF 199
Score = 29.6 bits (65), Expect = 2.8, Method: Composition-based stats.
Identities = 34/140 (24%), Positives = 62/140 (44%), Gaps = 8/140 (5%)
Query 1 LDRNIDNKALYDTFSLFGNILSCKVGVDEAN-RSKGYGFVHYESEASARNAIEKVNGMLI 59
LD +D + L + F GN+ + + D+ N GYGFV YE E A +N +
Sbjct 27 LDAQVDEEILCELFMQCGNVKNVHIPRDKINGYHAGYGFVEYEYEYECEYAANILNMTKL 86
Query 60 GGKTVYVGPFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVV--K 117
GK + ++ ++ + N+++ NL D + L I FG+I ++ V
Sbjct 87 FGKALRC----NKATQDKRSFDVGANLFIGNLDD-EVDEKMLFDIFSSFGQIMTVKVMRN 141
Query 118 QDPKGRPFAFCNYAEHSAAN 137
+D + F +Y +++
Sbjct 142 EDDTSKGHGFISYDNFESSD 161
> ath:AT1G73530 RNA recognition motif (RRM)-containing protein
Length=181
Score = 58.2 bits (139), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 32/95 (33%), Positives = 51/95 (53%), Gaps = 8/95 (8%)
Query 9 ALYDTFSLFGNILSCKVGVDE-ANRSKGYGFVHYESEASARNAIEKVNGMLIGGKTVYVG 67
L DTF FGN++ + +D+ ANR KG+ F+ YE+E A AI+ ++G + G+ ++V
Sbjct 92 TLRDTFEQFGNLIHMNMVMDKVANRPKGFAFLRYETEEEAMKAIQGMHGKFLDGRVIFVE 151
Query 68 PFITRSERESAGVNRFTNVYLKNLPRCWSDPESLR 102
TRS+ A R + P+ S P + R
Sbjct 152 EAKTRSDMSRAKPRR-------DFPKPQSKPRTFR 179
> dre:415211 puf60b, SIAHBP1, fe37c05, repressor, si:ch211-12p8.2,
si:zc12p8.2, wu:fb33e11, wu:fe37c05, zgc:86806; poly-U
binding splicing factor b
Length=502
Score = 58.2 bits (139), Expect = 8e-09, Method: Composition-based stats.
Identities = 38/130 (29%), Positives = 68/130 (52%), Gaps = 17/130 (13%)
Query 14 FSLFGNILSCKVGVDEAN-RSKGYGFVHYESEASARNAIEKVNGMLIGGKTVYVG----- 67
F+ FG I S + D + KG+ FV YE +A+ A+E++N +++GG+ + VG
Sbjct 89 FAPFGPIKSIDMSWDSVTMKHKGFAFVEYEVPEAAQLALEQMNSVMLGGRNIKVGRPGSI 148
Query 68 ----PFITRSERESAGVNRFTNVYLKNLPRCWSDPESLRPILEPFGEITSLVVKQDP--- 120
P I + E+ NR +Y+ ++ SD + ++ + E FG+I S ++ ++P
Sbjct 149 GQAQPIIEQLAEEARAYNR---IYVASIHPDLSD-DDIKSVFEAFGKIKSCMLAREPTTG 204
Query 121 KGRPFAFCNY 130
K + F F Y
Sbjct 205 KHKGFGFIEY 214
Lambda K H
0.318 0.136 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2487377096
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40