bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
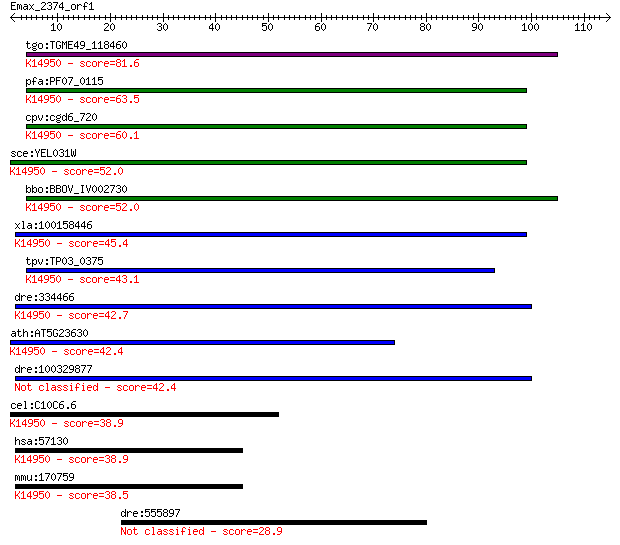
Query= Emax_2374_orf1
Length=114
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_118460 cation-transporting ATPase, putative (EC:3.6... 81.6 6e-16
pfa:PF07_0115 cation transporting ATPase, cation transporter; ... 63.5 1e-10
cpv:cgd6_720 hypothetical protein ; K14950 cation-transporting... 60.1 2e-09
sce:YEL031W SPF1, COD1, PER9, PIO1; P-type ATPase, ion transpo... 52.0 5e-07
bbo:BBOV_IV002730 21.m03018; cation transporting ATPase; K1495... 52.0 5e-07
xla:100158446 atp13a1; ATPase type 13A1 (EC:3.6.3.8); K14950 c... 45.4 4e-05
tpv:TP03_0375 integral membrane protein; K14950 cation-transpo... 43.1 2e-04
dre:334466 atp13a, hm:zehn0386, wu:fi75c10, zgc:63781; ATPase ... 42.7 3e-04
ath:AT5G23630 ATPase E1-E2 type family protein / haloacid deha... 42.4 4e-04
dre:100329877 ATPase type 13A-like 42.4 4e-04
cel:C10C6.6 hypothetical protein; K14950 cation-transporting A... 38.9 0.004
hsa:57130 ATP13A1, ATP13A, DKFZp761L1623, FLJ31858, FLJ41786, ... 38.9 0.005
mmu:170759 Atp13a1, Atp13a, Cgi152, catp; ATPase type 13A1 (EC... 38.5 0.005
dre:555897 transient receptor potential channel 4 alpha-like 28.9 4.0
> tgo:TGME49_118460 cation-transporting ATPase, putative (EC:3.6.3.2
3.6.3.8); K14950 cation-transporting ATPase 13A1 [EC:3.6.3.-]
Length=1484
Score = 81.6 bits (200), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 47/101 (46%), Positives = 65/101 (64%), Gaps = 0/101 (0%)
Query 4 FKPDAVNTATFYLLLAMHMSTFLANYEGAPWSRPLLQNARLGKMLLSGLVLLLVLLLQLL 63
F+P+ VN+ F L+ +MH STFL+NYEGAP+ PL +N L L + LL L+ +L+
Sbjct 1368 FEPNLVNSVVFLLIASMHASTFLSNYEGAPFMVPLTENKPLVLTLGFLVSTLLTLVFELV 1427
Query 64 PFLNEILDLVLFHDADFLLQLQLLVAADLLGPWLVAATCRK 104
P LNE L LV F +F +++ LV DL+G WLVA + RK
Sbjct 1428 PSLNETLSLVPFPTREFKVKIIGLVFLDLVGAWLVAWSLRK 1468
> pfa:PF07_0115 cation transporting ATPase, cation transporter;
K14950 cation-transporting ATPase 13A1 [EC:3.6.3.-]
Length=1918
Score = 63.5 bits (153), Expect = 1e-10, Method: Composition-based stats.
Identities = 32/95 (33%), Positives = 50/95 (52%), Gaps = 0/95 (0%)
Query 4 FKPDAVNTATFYLLLAMHMSTFLANYEGAPWSRPLLQNARLGKMLLSGLVLLLVLLLQLL 63
F P+ VNT +YL+ +++S F NYEG P+ P+ +N + + L VL++ +
Sbjct 1805 FIPNLVNTCIYYLIYCINLSIFSCNYEGLPFMVPIHKNKEIVYIFAVNFFFLFVLVMDIF 1864
Query 64 PFLNEILDLVLFHDADFLLQLQLLVAADLLGPWLV 98
PFLN LV F + F L+ D+ P+LV
Sbjct 1865 PFLNYFFSLVSFPNIRFKFFFFFLMLVDIFLPYLV 1899
> cpv:cgd6_720 hypothetical protein ; K14950 cation-transporting
ATPase 13A1 [EC:3.6.3.-]
Length=1088
Score = 60.1 bits (144), Expect = 2e-09, Method: Composition-based stats.
Identities = 34/97 (35%), Positives = 54/97 (55%), Gaps = 4/97 (4%)
Query 4 FKPDAVNTATFYLLLAMHMSTFLANYEGAPWSRPLLQNARLGKMLLSGLVL--LLVLLLQ 61
F+P+ +NT +YL A H++ FL+N +G P++ PL +N L + S LV+ L+ +L
Sbjct 978 FEPNIINTTMYYLYTACHLACFLSNAQGHPFTTPLSENKYL--IYTSALVISFLITSILG 1035
Query 62 LLPFLNEILDLVLFHDADFLLQLQLLVAADLLGPWLV 98
+ P LN + LV + L LLV D+ G L+
Sbjct 1036 IFPHLNILFSLVTLQSKYYQSILILLVLFDIFGTLLI 1072
> sce:YEL031W SPF1, COD1, PER9, PIO1; P-type ATPase, ion transporter
of the ER membrane involved in ER function and Ca2+ homeostasis;
required for regulating Hmg2p degradation; confers
sensitivity to a killer toxin (SMKT) produced by Pichia farinosa
KK1 (EC:3.6.3.-); K14950 cation-transporting ATPase
13A1 [EC:3.6.3.-]
Length=1215
Score = 52.0 bits (123), Expect = 5e-07, Method: Composition-based stats.
Identities = 34/98 (34%), Positives = 47/98 (47%), Gaps = 1/98 (1%)
Query 1 EEAFKPDAVNTATFYLLLAMHMSTFLANYEGAPWSRPLLQNARLGKMLLSGLVLLLVLLL 60
E+ F P +NT F + L +STF NY+G P+ + N + LL L L
Sbjct 1093 EKEFAPSLLNTGIFIIQLVQQVSTFAVNYQGEPFRENIRSNKGMYYGLLGVTGLALASAT 1152
Query 61 QLLPFLNEILDLVLFHDADFLLQLQLLVAADLLGPWLV 98
+ LP LNE + V D DF ++L L + D G W V
Sbjct 1153 EFLPELNEAMKFVPMTD-DFKIKLTLTLLLDFFGSWGV 1189
> bbo:BBOV_IV002730 21.m03018; cation transporting ATPase; K14950
cation-transporting ATPase 13A1 [EC:3.6.3.-]
Length=1246
Score = 52.0 bits (123), Expect = 5e-07, Method: Composition-based stats.
Identities = 28/101 (27%), Positives = 51/101 (50%), Gaps = 0/101 (0%)
Query 4 FKPDAVNTATFYLLLAMHMSTFLANYEGAPWSRPLLQNARLGKMLLSGLVLLLVLLLQLL 63
F+ + VNT FY +++S FLANY+G P+ L+ N + + L + +L+L+ L+
Sbjct 1142 FEANLVNTLVFYGCFVVNISGFLANYQGYPYMESLMDNKFVYRPLAAATFFILLLVCDLI 1201
Query 64 PFLNEILDLVLFHDADFLLQLQLLVAADLLGPWLVAATCRK 104
L LV L + ++ D++G + + CR+
Sbjct 1202 SPLANFFSLVEIPGHMIRLHVIAILLLDMVGSFFIEHGCRR 1242
> xla:100158446 atp13a1; ATPase type 13A1 (EC:3.6.3.8); K14950
cation-transporting ATPase 13A1 [EC:3.6.3.-]
Length=1174
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 32/102 (31%), Positives = 52/102 (50%), Gaps = 11/102 (10%)
Query 2 EAFKPDAVNTATFYLLLAMHMSTFLANYEGAPWSRPLLQNAR-LGKMLLSGLVLLLVLLL 60
+ F+P VN+ + + +AM M+TF NY+G P+ L +N L ++LSG V +L LL
Sbjct 1062 KEFEPSLVNSTVYIMSMAMQMATFAINYKGHPFMESLRENKPLLWSIILSG-VAILGLLS 1120
Query 61 QLLPFLNEILDLVLFHDADFLLQLQLLVA----ADLLGPWLV 98
P N+ LV D ++ ++++A D W V
Sbjct 1121 GSSPEFNDQFSLV-----DIPMEFKIVIAKVLFVDFCTAWAV 1157
> tpv:TP03_0375 integral membrane protein; K14950 cation-transporting
ATPase 13A1 [EC:3.6.3.-]
Length=1522
Score = 43.1 bits (100), Expect = 2e-04, Method: Composition-based stats.
Identities = 27/89 (30%), Positives = 46/89 (51%), Gaps = 1/89 (1%)
Query 4 FKPDAVNTATFYLLLAMHMSTFLANYEGAPWSRPLLQNARLGKMLLSGLVLLLVLLLQLL 63
F+P+ VNT +Y+ A+ +++F++NY P+ + N L K +L LL+ + L+
Sbjct 1417 FEPNVVNTLLYYVWYAISLNSFVSNYVDYPYMDTISNNKFLFKPILLSYFTLLLFISDLV 1476
Query 64 PFLNEILDLVLFHDADFLLQLQLLVAADL 92
LN LV L++ L+V DL
Sbjct 1477 KPLNSFFSLVPISHM-LKLKISLVVLFDL 1504
> dre:334466 atp13a, hm:zehn0386, wu:fi75c10, zgc:63781; ATPase
type 13A; K14950 cation-transporting ATPase 13A1 [EC:3.6.3.-]
Length=1177
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 30/99 (30%), Positives = 52/99 (52%), Gaps = 7/99 (7%)
Query 2 EAFKPDAVNTATFYLLLAMHMSTFLANYEGAPWSRPLLQNAR-LGKMLLSGLVLLLVLLL 60
+ F+P +N+ + + +AM M+TF NY+G P+ L +N L + +SGL ++ LL
Sbjct 1065 KEFEPSLINSTVYIMSMAMQMATFAINYKGHPFMESLTENRPLLWSIAISGLA-IVGLLT 1123
Query 61 QLLPFLNEILDLVLFHDADFLLQLQLLVAADLLGPWLVA 99
P NE LV D + +L++A L+ ++ A
Sbjct 1124 GSSPEFNEQFALV-----DIPTEFKLIIAQVLVVDFVAA 1157
> ath:AT5G23630 ATPase E1-E2 type family protein / haloacid dehalogenase-like
hydrolase familiy protein; K14950 cation-transporting
ATPase 13A1 [EC:3.6.3.-]
Length=1179
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 40/73 (54%), Gaps = 0/73 (0%)
Query 1 EEAFKPDAVNTATFYLLLAMHMSTFLANYEGAPWSRPLLQNARLGKMLLSGLVLLLVLLL 60
+ +F P+ VNT ++ + + + ++TF NY G P+++ + +N L++G V+
Sbjct 1051 DASFHPNLVNTVSYMVSMMLQVATFAVNYMGHPFNQSIRENKPFFYALIAGAGFFTVIAS 1110
Query 61 QLLPFLNEILDLV 73
L LN+ L LV
Sbjct 1111 DLFRDLNDSLKLV 1123
> dre:100329877 ATPase type 13A-like
Length=391
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 30/99 (30%), Positives = 52/99 (52%), Gaps = 7/99 (7%)
Query 2 EAFKPDAVNTATFYLLLAMHMSTFLANYEGAPWSRPLLQNAR-LGKMLLSGLVLLLVLLL 60
+ F+P +N+ + + +AM M+TF NY+G P+ L +N L + +SGL ++ LL
Sbjct 279 KEFEPSLINSTVYIMSMAMQMATFAINYKGHPFMESLTENRPLLWSIAISGLA-IVGLLT 337
Query 61 QLLPFLNEILDLVLFHDADFLLQLQLLVAADLLGPWLVA 99
P NE LV D + +L++A L+ ++ A
Sbjct 338 GSSPEFNEQFALV-----DIPTEFKLIIAQVLVVDFVAA 371
> cel:C10C6.6 hypothetical protein; K14950 cation-transporting
ATPase 13A1 [EC:3.6.3.-]
Length=1178
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 29/52 (55%), Gaps = 1/52 (1%)
Query 1 EEAFKPDAVNTATFYLLLAMHMSTFLANYEGAPWSRPLLQN-ARLGKMLLSG 51
E F P+ +NT + + +A+ + TF NY G P+ L +N A L ++ SG
Sbjct 1070 EAKFTPNILNTTVYIISMALQVCTFAVNYRGRPFMESLFENKAMLYSIMFSG 1121
> hsa:57130 ATP13A1, ATP13A, DKFZp761L1623, FLJ31858, FLJ41786,
FLJ43873, FLJ90317, KIAA1825; ATPase type 13A1 (EC:3.6.3.8
3.6.1.38); K14950 cation-transporting ATPase 13A1 [EC:3.6.3.-]
Length=1204
Score = 38.9 bits (89), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 26/43 (60%), Gaps = 0/43 (0%)
Query 2 EAFKPDAVNTATFYLLLAMHMSTFLANYEGAPWSRPLLQNARL 44
+ F+P VN+ + + +AM M+TF NY+G P+ L +N L
Sbjct 1092 KEFEPSLVNSTVYIMAMAMQMATFAINYKGPPFMESLPENKPL 1134
> mmu:170759 Atp13a1, Atp13a, Cgi152, catp; ATPase type 13A1 (EC:3.6.3.8);
K14950 cation-transporting ATPase 13A1 [EC:3.6.3.-]
Length=1200
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 26/43 (60%), Gaps = 0/43 (0%)
Query 2 EAFKPDAVNTATFYLLLAMHMSTFLANYEGAPWSRPLLQNARL 44
+ F+P VN+ + + +AM M+TF NY+G P+ L +N L
Sbjct 1088 KEFEPSLVNSTVYIMAMAMQMATFAINYKGPPFMESLPENKPL 1130
> dre:555897 transient receptor potential channel 4 alpha-like
Length=934
Score = 28.9 bits (63), Expect = 4.0, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 29/58 (50%), Gaps = 6/58 (10%)
Query 22 MSTFLANYEGAPWSRPLLQNARLGKMLLSGLVLLLVLLLQLLPFLNEILDLVLFHDAD 79
+S F AN P LQ LG+MLL L L + L LL F N + L ++++D
Sbjct 504 ISLFTANSHLGP-----LQ-ISLGRMLLDILKFLFIYCLVLLAFANGLNQLYFYYESD 555
Lambda K H
0.328 0.142 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2047611720
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40