bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2364_orf1
Length=76
Score E
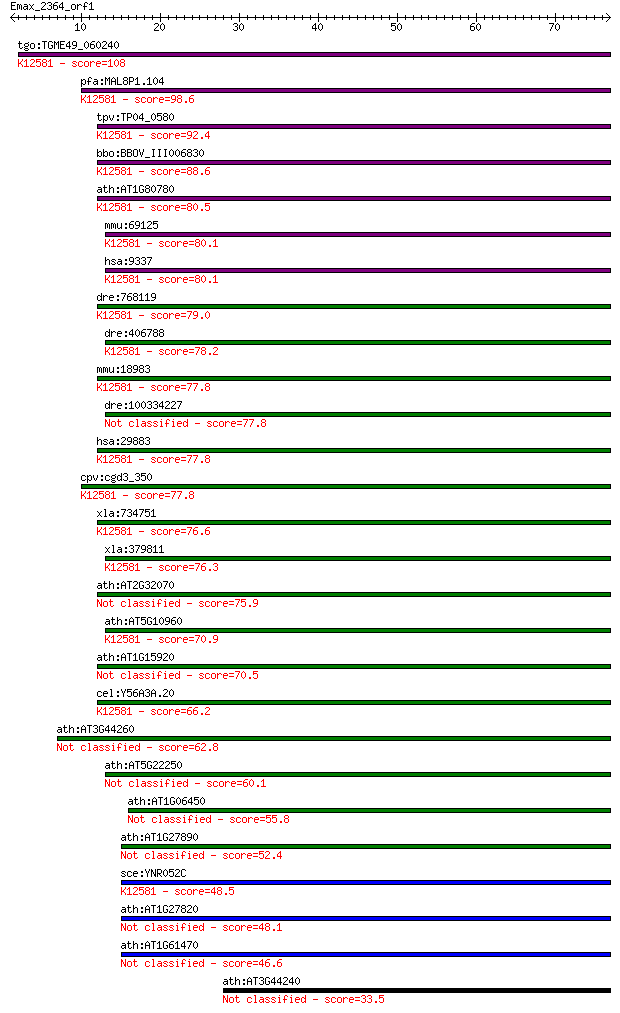
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_060240 CCR4-NOT transcription complex subunit, puta... 108 4e-24
pfa:MAL8P1.104 CAF1 family ribonuclease, putative; K12581 CCR4... 98.6 4e-21
tpv:TP04_0580 hypothetical protein; K12581 CCR4-NOT transcript... 92.4 3e-19
bbo:BBOV_III006830 17.m07605; CAF1 family ribonuclease contain... 88.6 5e-18
ath:AT1G80780 CCR4-NOT transcription complex protein, putative... 80.5 1e-15
mmu:69125 Cnot8, 1500015I04Rik, 1810022F04Rik, AA536816, AU015... 80.1 2e-15
hsa:9337 CNOT8, CAF1, CALIF, POP2, hCAF1; CCR4-NOT transcripti... 80.1 2e-15
dre:768119 cnot7, CAF1, fd59c07, fj07d05, wu:fd59c07, wu:fj07d... 79.0 3e-15
dre:406788 cnot8, wu:fe49a05, zgc:63844; CCR4-NOT transcriptio... 78.2 7e-15
mmu:18983 Cnot7, AU022737, Caf1, Pop2; CCR4-NOT transcription ... 77.8 7e-15
dre:100334227 CCR4-NOT transcription complex, subunit 8-like 77.8 7e-15
hsa:29883 CNOT7, CAF1, hCAF-1; CCR4-NOT transcription complex,... 77.8 8e-15
cpv:cgd3_350 Pop2p-like 3'5' exonuclease, CCR4-NOT transcripti... 77.8 9e-15
xla:734751 cnot7, MGC130876, caf1; CCR4-NOT transcription comp... 76.6 2e-14
xla:379811 cnot8, MGC52767; CCR4-NOT transcription complex, su... 76.3 2e-14
ath:AT2G32070 CCR4-NOT transcription complex protein, putative 75.9 3e-14
ath:AT5G10960 CCR4-NOT transcription complex protein, putative... 70.9 1e-12
ath:AT1G15920 CCR4-NOT transcription complex protein, putative 70.5 1e-12
cel:Y56A3A.20 ccf-1; yeast CCR4 Associated Factor family membe... 66.2 2e-11
ath:AT3G44260 CCR4-NOT transcription complex protein, putative 62.8 3e-10
ath:AT5G22250 CCR4-NOT transcription complex protein, putative 60.1 2e-09
ath:AT1G06450 CCR4-NOT transcription complex protein, putative 55.8 3e-08
ath:AT1G27890 CCR4-NOT transcription complex protein, putative 52.4 4e-07
sce:YNR052C POP2, CAF1; Pop2p (EC:3.1.13.4); K12581 CCR4-NOT t... 48.5 5e-06
ath:AT1G27820 CCR4-NOT transcription complex protein, putative 48.1 7e-06
ath:AT1G61470 CCR4-NOT transcription complex protein, putative 46.6 2e-05
ath:AT3G44240 CCR4-NOT transcription complex protein, putative 33.5 0.17
> tgo:TGME49_060240 CCR4-NOT transcription complex subunit, putative
(EC:3.1.13.4); K12581 CCR4-NOT transcription complex
subunit 7/8
Length=617
Score = 108 bits (270), Expect = 4e-24, Method: Composition-based stats.
Identities = 54/75 (72%), Positives = 58/75 (77%), Gaps = 3/75 (4%)
Query 2 MNGYGDGLRGQIIEVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSMLNALDYN 61
MNG G R QI+EVW HNLEEE RI DVVER+ YIA+DTEFPGIV RPT N DYN
Sbjct 1 MNGEC-GEREQIVEVWEHNLEEEFARIRDVVERFQYIAMDTEFPGIVARPTG--NVTDYN 57
Query 62 YRIVKYNVDLLKVIQ 76
Y+ VKYNVDLLKVIQ
Sbjct 58 YQTVKYNVDLLKVIQ 72
> pfa:MAL8P1.104 CAF1 family ribonuclease, putative; K12581 CCR4-NOT
transcription complex subunit 7/8
Length=1774
Score = 98.6 bits (244), Expect = 4e-21, Method: Composition-based stats.
Identities = 45/67 (67%), Positives = 57/67 (85%), Gaps = 2/67 (2%)
Query 10 RGQIIEVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSMLNALDYNYRIVKYNV 69
R +I++VWA+NLEEE +RI D+VE++PY+A+DTEFPGIV RPT N LDYNY+ +K NV
Sbjct 4 RTKIVDVWANNLEEEFERIRDIVEKHPYVAIDTEFPGIVARPTG--NVLDYNYQTIKCNV 61
Query 70 DLLKVIQ 76
DLLKVIQ
Sbjct 62 DLLKVIQ 68
> tpv:TP04_0580 hypothetical protein; K12581 CCR4-NOT transcription
complex subunit 7/8
Length=562
Score = 92.4 bits (228), Expect = 3e-19, Method: Composition-based stats.
Identities = 41/65 (63%), Positives = 53/65 (81%), Gaps = 2/65 (3%)
Query 12 QIIEVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSMLNALDYNYRIVKYNVDL 71
QI++VW+ NLE+ DRI D++E+YPY+++DTEFPGIV RPTS L DYNY+ VK NVDL
Sbjct 6 QIVDVWSDNLEDAFDRIRDLLEQYPYVSIDTEFPGIVVRPTSYLE--DYNYQTVKCNVDL 63
Query 72 LKVIQ 76
L +IQ
Sbjct 64 LNIIQ 68
> bbo:BBOV_III006830 17.m07605; CAF1 family ribonuclease containing
protein; K12581 CCR4-NOT transcription complex subunit
7/8
Length=374
Score = 88.6 bits (218), Expect = 5e-18, Method: Composition-based stats.
Identities = 39/65 (60%), Positives = 53/65 (81%), Gaps = 2/65 (3%)
Query 12 QIIEVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSMLNALDYNYRIVKYNVDL 71
+I++VW+ NLE+ +RI DV+ERYPY+++DTEFPGIV +PT+ DYNY+ VK NVDL
Sbjct 6 KIVDVWSDNLEDAFERIRDVLERYPYVSIDTEFPGIVAKPTTYQE--DYNYQTVKCNVDL 63
Query 72 LKVIQ 76
LK+IQ
Sbjct 64 LKLIQ 68
> ath:AT1G80780 CCR4-NOT transcription complex protein, putative;
K12581 CCR4-NOT transcription complex subunit 7/8
Length=274
Score = 80.5 bits (197), Expect = 1e-15, Method: Composition-based stats.
Identities = 37/66 (56%), Positives = 49/66 (74%), Gaps = 1/66 (1%)
Query 12 QIIEVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSMLNA-LDYNYRIVKYNVD 70
QI EVW NL+EE+D I DVV+ +PY+A+DTEFPGIV RP + DY+Y +K NV+
Sbjct 11 QIREVWNDNLQEEMDLIRDVVDDFPYVAMDTEFPGIVVRPVGTFKSNADYHYETLKTNVN 70
Query 71 LLKVIQ 76
+LK+IQ
Sbjct 71 ILKMIQ 76
> mmu:69125 Cnot8, 1500015I04Rik, 1810022F04Rik, AA536816, AU015770,
AU043059; CCR4-NOT transcription complex, subunit 8;
K12581 CCR4-NOT transcription complex subunit 7/8
Length=292
Score = 80.1 bits (196), Expect = 2e-15, Method: Composition-based stats.
Identities = 36/65 (55%), Positives = 50/65 (76%), Gaps = 1/65 (1%)
Query 13 IIEVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSML-NALDYNYRIVKYNVDL 71
I EVWA NLEEE+ +I ++V Y YIA+DTEFPG+V RP +++DY Y++++ NVDL
Sbjct 12 ICEVWASNLEEEMRKIREIVLSYSYIAMDTEFPGVVVRPIGEFRSSIDYQYQLLRCNVDL 71
Query 72 LKVIQ 76
LK+IQ
Sbjct 72 LKIIQ 76
> hsa:9337 CNOT8, CAF1, CALIF, POP2, hCAF1; CCR4-NOT transcription
complex, subunit 8; K12581 CCR4-NOT transcription complex
subunit 7/8
Length=292
Score = 80.1 bits (196), Expect = 2e-15, Method: Composition-based stats.
Identities = 36/65 (55%), Positives = 50/65 (76%), Gaps = 1/65 (1%)
Query 13 IIEVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSML-NALDYNYRIVKYNVDL 71
I EVWA NLEEE+ +I ++V Y YIA+DTEFPG+V RP +++DY Y++++ NVDL
Sbjct 12 ICEVWASNLEEEMRKIREIVLSYSYIAMDTEFPGVVVRPIGEFRSSIDYQYQLLRCNVDL 71
Query 72 LKVIQ 76
LK+IQ
Sbjct 72 LKIIQ 76
> dre:768119 cnot7, CAF1, fd59c07, fj07d05, wu:fd59c07, wu:fj07d05,
zgc:153168; CCR4-NOT transcription complex, subunit 7;
K12581 CCR4-NOT transcription complex subunit 7/8
Length=286
Score = 79.0 bits (193), Expect = 3e-15, Method: Composition-based stats.
Identities = 36/66 (54%), Positives = 48/66 (72%), Gaps = 1/66 (1%)
Query 12 QIIEVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSMLNA-LDYNYRIVKYNVD 70
+I EVWA NLEEE+ RI V ++ YIA+DTEFPG+V RP + DY Y++++ NVD
Sbjct 11 RICEVWACNLEEEMKRIRQVTRKFNYIAMDTEFPGVVARPIGEFRSNADYQYQLLRCNVD 70
Query 71 LLKVIQ 76
LLK+IQ
Sbjct 71 LLKIIQ 76
> dre:406788 cnot8, wu:fe49a05, zgc:63844; CCR4-NOT transcription
complex, subunit 8; K12581 CCR4-NOT transcription complex
subunit 7/8
Length=285
Score = 78.2 bits (191), Expect = 7e-15, Method: Composition-based stats.
Identities = 33/65 (50%), Positives = 49/65 (75%), Gaps = 1/65 (1%)
Query 13 IIEVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSMLNA-LDYNYRIVKYNVDL 71
I EVWA N+EEE+ +I +++ Y YIA+DTEFPG+V RP + +DY Y++++ NVDL
Sbjct 12 ICEVWASNVEEEMRKIRQIIQNYNYIAMDTEFPGVVVRPIGEFRSTVDYQYQLLRCNVDL 71
Query 72 LKVIQ 76
LK++Q
Sbjct 72 LKIVQ 76
> mmu:18983 Cnot7, AU022737, Caf1, Pop2; CCR4-NOT transcription
complex, subunit 7; K12581 CCR4-NOT transcription complex
subunit 7/8
Length=285
Score = 77.8 bits (190), Expect = 7e-15, Method: Composition-based stats.
Identities = 34/66 (51%), Positives = 49/66 (74%), Gaps = 1/66 (1%)
Query 12 QIIEVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSMLNA-LDYNYRIVKYNVD 70
+I EVWA NL+EE+ +I V+ +Y Y+A+DTEFPG+V RP + DY Y++++ NVD
Sbjct 11 RICEVWACNLDEEMKKIRQVIRKYNYVAMDTEFPGVVARPIGEFRSNADYQYQLLRCNVD 70
Query 71 LLKVIQ 76
LLK+IQ
Sbjct 71 LLKIIQ 76
> dre:100334227 CCR4-NOT transcription complex, subunit 8-like
Length=247
Score = 77.8 bits (190), Expect = 7e-15, Method: Composition-based stats.
Identities = 34/65 (52%), Positives = 49/65 (75%), Gaps = 1/65 (1%)
Query 13 IIEVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSMLNA-LDYNYRIVKYNVDL 71
I EVWA N+EEE+ +I +++ Y YIA+DTEFPG+V RP + +DY Y++++ NVDL
Sbjct 12 ICEVWASNVEEEMRKIRQIIQNYNYIAMDTEFPGVVVRPIGEFRSTVDYQYQLLRCNVDL 71
Query 72 LKVIQ 76
LK+IQ
Sbjct 72 LKIIQ 76
> hsa:29883 CNOT7, CAF1, hCAF-1; CCR4-NOT transcription complex,
subunit 7; K12581 CCR4-NOT transcription complex subunit
7/8
Length=285
Score = 77.8 bits (190), Expect = 8e-15, Method: Composition-based stats.
Identities = 34/66 (51%), Positives = 49/66 (74%), Gaps = 1/66 (1%)
Query 12 QIIEVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSMLNA-LDYNYRIVKYNVD 70
+I EVWA NL+EE+ +I V+ +Y Y+A+DTEFPG+V RP + DY Y++++ NVD
Sbjct 11 RICEVWACNLDEEMKKIRQVIRKYNYVAMDTEFPGVVARPIGEFRSNADYQYQLLRCNVD 70
Query 71 LLKVIQ 76
LLK+IQ
Sbjct 71 LLKIIQ 76
> cpv:cgd3_350 Pop2p-like 3'5' exonuclease, CCR4-NOT transcription
complex ; K12581 CCR4-NOT transcription complex subunit
7/8
Length=277
Score = 77.8 bits (190), Expect = 9e-15, Method: Composition-based stats.
Identities = 34/67 (50%), Positives = 50/67 (74%), Gaps = 2/67 (2%)
Query 10 RGQIIEVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSMLNALDYNYRIVKYNV 69
+G I EVW +N+ E I ++++ +PY+A+DTEFPG+V RPT+ N +Y Y+ V++NV
Sbjct 15 KGVIYEVWQNNINEAFQMISEIMDDFPYVAIDTEFPGVVVRPTN--NYYEYYYQTVRFNV 72
Query 70 DLLKVIQ 76
DLLKVIQ
Sbjct 73 DLLKVIQ 79
> xla:734751 cnot7, MGC130876, caf1; CCR4-NOT transcription complex,
subunit 7; K12581 CCR4-NOT transcription complex subunit
7/8
Length=285
Score = 76.6 bits (187), Expect = 2e-14, Method: Composition-based stats.
Identities = 33/66 (50%), Positives = 49/66 (74%), Gaps = 1/66 (1%)
Query 12 QIIEVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSMLNA-LDYNYRIVKYNVD 70
+I EVWA NL++++ RI V+ +Y Y+A+DTEFPG+V RP + DY Y++++ NVD
Sbjct 11 RICEVWACNLDDQMKRIRQVIRKYNYVAMDTEFPGVVARPIGEFRSNADYQYQLLRCNVD 70
Query 71 LLKVIQ 76
LLK+IQ
Sbjct 71 LLKIIQ 76
> xla:379811 cnot8, MGC52767; CCR4-NOT transcription complex,
subunit 8; K12581 CCR4-NOT transcription complex subunit 7/8
Length=289
Score = 76.3 bits (186), Expect = 2e-14, Method: Composition-based stats.
Identities = 35/65 (53%), Positives = 49/65 (75%), Gaps = 1/65 (1%)
Query 13 IIEVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSMLNA-LDYNYRIVKYNVDL 71
I EVWA NLEEE+ +I ++V + YIA+DTEFPG+V RP + +DY Y++++ NVDL
Sbjct 12 ICEVWAVNLEEEMRKIRELVRTHGYIAMDTEFPGVVVRPIGEFRSTIDYQYQLLRCNVDL 71
Query 72 LKVIQ 76
LK+IQ
Sbjct 72 LKIIQ 76
> ath:AT2G32070 CCR4-NOT transcription complex protein, putative
Length=275
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 34/66 (51%), Positives = 47/66 (71%), Gaps = 1/66 (1%)
Query 12 QIIEVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSMLNA-LDYNYRIVKYNVD 70
QI EVW NLE E+ I +VV+ +P++A+DTEFPGIVCRP +Y+Y +K NV+
Sbjct 11 QIREVWNDNLESEMALIREVVDDFPFVAMDTEFPGIVCRPVGTFKTNTEYHYETLKTNVN 70
Query 71 LLKVIQ 76
+LK+IQ
Sbjct 71 ILKMIQ 76
> ath:AT5G10960 CCR4-NOT transcription complex protein, putative;
K12581 CCR4-NOT transcription complex subunit 7/8
Length=277
Score = 70.9 bits (172), Expect = 1e-12, Method: Composition-based stats.
Identities = 34/65 (52%), Positives = 46/65 (70%), Gaps = 1/65 (1%)
Query 13 IIEVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSMLN-ALDYNYRIVKYNVDL 71
I EVW +NL EE I ++V+++ YIA+DTEFPG+V +P + D NYR +K NVDL
Sbjct 12 IREVWDYNLVEEFALIREIVDKFSYIAMDTEFPGVVLKPVATFKYNNDLNYRTLKENVDL 71
Query 72 LKVIQ 76
LK+IQ
Sbjct 72 LKLIQ 76
> ath:AT1G15920 CCR4-NOT transcription complex protein, putative
Length=286
Score = 70.5 bits (171), Expect = 1e-12, Method: Composition-based stats.
Identities = 33/72 (45%), Positives = 47/72 (65%), Gaps = 7/72 (9%)
Query 12 QIIEVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCR-------PTSMLNALDYNYRI 64
+I EVW HNLE+E+ I ++ +PY+A+DTEFPGIVC+ P +YNY
Sbjct 15 EIREVWNHNLEQEMALIEQSIDDFPYVAMDTEFPGIVCKTVTANPNPNPYSIHYEYNYDT 74
Query 65 VKYNVDLLKVIQ 76
+K NV++LK+IQ
Sbjct 75 LKANVNMLKLIQ 86
> cel:Y56A3A.20 ccf-1; yeast CCR4 Associated Factor family member
(ccf-1); K12581 CCR4-NOT transcription complex subunit 7/8
Length=310
Score = 66.2 bits (160), Expect = 2e-11, Method: Composition-based stats.
Identities = 32/66 (48%), Positives = 44/66 (66%), Gaps = 1/66 (1%)
Query 12 QIIEVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSMLNAL-DYNYRIVKYNVD 70
+I V+ N+EEE RI VE YPY+A+DTEFPG+V P + D+NY+ V NV+
Sbjct 22 KIHNVYMSNVEEEFARIRGFVEDYPYVAMDTEFPGVVATPLGTFRSKEDFNYQQVFCNVN 81
Query 71 LLKVIQ 76
+LK+IQ
Sbjct 82 MLKLIQ 87
> ath:AT3G44260 CCR4-NOT transcription complex protein, putative
Length=280
Score = 62.8 bits (151), Expect = 3e-10, Method: Composition-based stats.
Identities = 29/70 (41%), Positives = 44/70 (62%), Gaps = 0/70 (0%)
Query 7 DGLRGQIIEVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSMLNALDYNYRIVK 66
DG+ EVWA NLE E + I ++++ YP+I++DTEFPG++ + D Y ++K
Sbjct 13 DGVTVVTREVWAENLESEFELISEIIDDYPFISMDTEFPGVIFKSDLRFTNPDDLYTLLK 72
Query 67 YNVDLLKVIQ 76
NVD L +IQ
Sbjct 73 ANVDALSLIQ 82
> ath:AT5G22250 CCR4-NOT transcription complex protein, putative
Length=278
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 32/69 (46%), Positives = 46/69 (66%), Gaps = 7/69 (10%)
Query 13 IIEVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSMLNAL-----DYNYRIVKY 67
I +VWA+NLE E D I +VE YP+I++DTEFPG++ + L+ L +Y Y ++K
Sbjct 14 IRDVWAYNLESEFDLIRGIVEDYPFISMDTEFPGVIYKAD--LDVLRRGNPNYLYNLLKS 71
Query 68 NVDLLKVIQ 76
NVD L +IQ
Sbjct 72 NVDALSLIQ 80
> ath:AT1G06450 CCR4-NOT transcription complex protein, putative
Length=360
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 27/61 (44%), Positives = 42/61 (68%), Gaps = 1/61 (1%)
Query 16 VWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSMLNALDYNYRIVKYNVDLLKVI 75
VW N++EE+ R+ + ++R+P IA DTE+PGI+ R T ++ D YR +K NV+ K+I
Sbjct 12 VWRSNVDEEMARMAECLKRFPLIAFDTEYPGIIFR-TYFDSSSDECYRAMKGNVENTKLI 70
Query 76 Q 76
Q
Sbjct 71 Q 71
> ath:AT1G27890 CCR4-NOT transcription complex protein, putative
Length=302
Score = 52.4 bits (124), Expect = 4e-07, Method: Composition-based stats.
Identities = 27/62 (43%), Positives = 39/62 (62%), Gaps = 1/62 (1%)
Query 15 EVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSMLNALDYNYRIVKYNVDLLKV 74
EVW N E E+D I D ++ + IA+DTEFPG + + T M + + YR +K+NVD +
Sbjct 3 EVWRWNKEVEMDSIRDCLKHFSSIAIDTEFPGCL-KETPMDASEEIRYRDMKFNVDNTHL 61
Query 75 IQ 76
IQ
Sbjct 62 IQ 63
> sce:YNR052C POP2, CAF1; Pop2p (EC:3.1.13.4); K12581 CCR4-NOT
transcription complex subunit 7/8
Length=433
Score = 48.5 bits (114), Expect = 5e-06, Method: Composition-based stats.
Identities = 23/63 (36%), Positives = 37/63 (58%), Gaps = 1/63 (1%)
Query 15 EVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSMLNA-LDYNYRIVKYNVDLLK 73
+VW NL E I +V +Y ++++ TEF G + RP + +DY+Y+ ++ NVD L
Sbjct 162 DVWKSNLYSEFAVIRQLVSQYNHVSISTEFVGTLARPIGTFRSKVDYHYQTMRANVDFLN 221
Query 74 VIQ 76
IQ
Sbjct 222 PIQ 224
> ath:AT1G27820 CCR4-NOT transcription complex protein, putative
Length=310
Score = 48.1 bits (113), Expect = 7e-06, Method: Composition-based stats.
Identities = 26/62 (41%), Positives = 38/62 (61%), Gaps = 1/62 (1%)
Query 15 EVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSMLNALDYNYRIVKYNVDLLKV 74
EVW N E E++ I D ++ IA+DTEFPG + + T M + + YR +K+NVD +
Sbjct 8 EVWRWNKEVEMNSIRDCLKHCSSIAIDTEFPGCL-KETPMDASEEIRYRDMKFNVDNTHL 66
Query 75 IQ 76
IQ
Sbjct 67 IQ 68
> ath:AT1G61470 CCR4-NOT transcription complex protein, putative
Length=278
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 25/62 (40%), Positives = 38/62 (61%), Gaps = 1/62 (1%)
Query 15 EVWAHNLEEEVDRIMDVVERYPYIALDTEFPGIVCRPTSMLNALDYNYRIVKYNVDLLKV 74
EVW N + E++ I D ++ IA+DTEFPG + + T M + + YR +K+NVD +
Sbjct 4 EVWRWNKQAEMNSIRDCLKHCNSIAIDTEFPGCL-KETPMDASDEIRYRDMKFNVDNTHL 62
Query 75 IQ 76
IQ
Sbjct 63 IQ 64
> ath:AT3G44240 CCR4-NOT transcription complex protein, putative
Length=239
Score = 33.5 bits (75), Expect = 0.17, Method: Composition-based stats.
Identities = 18/49 (36%), Positives = 28/49 (57%), Gaps = 1/49 (2%)
Query 28 IMDVVERYPYIALDTEFPGIVCRPTSMLNALDYNYRIVKYNVDLLKVIQ 76
I D + Y +IA+DTEFP + R T+ + Y + ++VD K+IQ
Sbjct 4 IEDCLRSYRFIAIDTEFPSTL-RETTQHATDEERYMDMSFSVDRAKLIQ 51
Lambda K H
0.323 0.144 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2076916240
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40