bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2302_orf1
Length=78
Score E
Sequences producing significant alignments: (Bits) Value
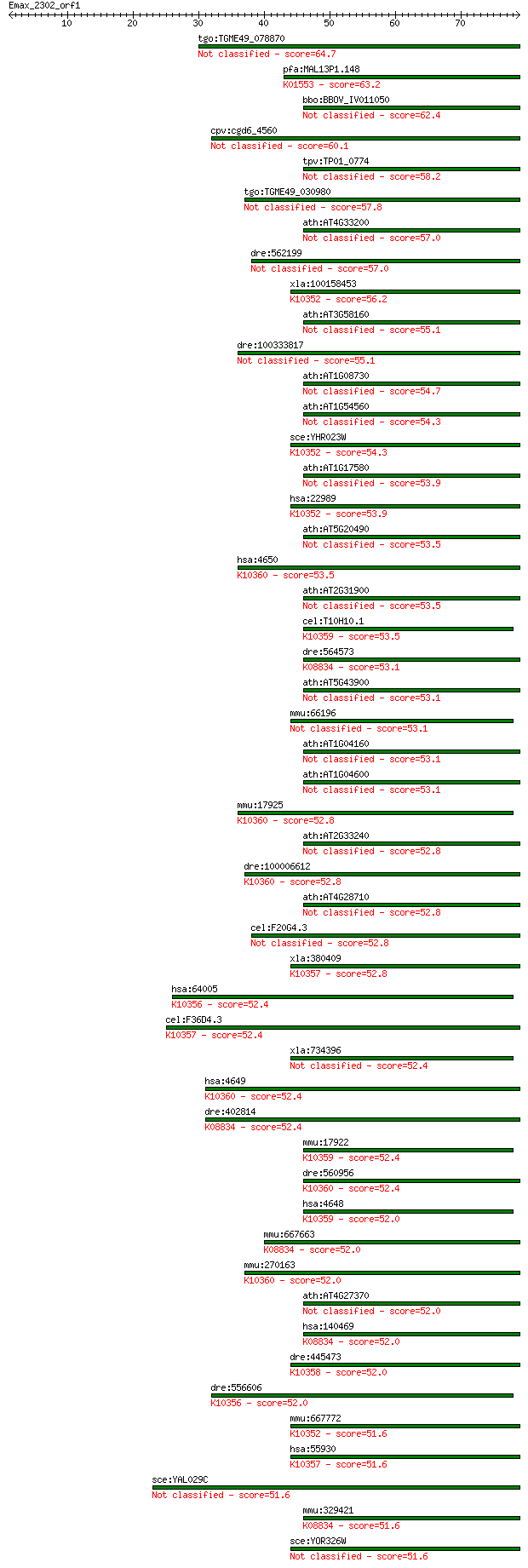
tgo:TGME49_078870 myosin F (TgMyoF) protein (EC:2.7.11.1 1.14.... 64.7 7e-11
pfa:MAL13P1.148 myosin; K01553 myosin ATPase [EC:3.6.4.1] 63.2 2e-10
bbo:BBOV_IV011050 23.m05995; WD40 repeat myosin-like protein (... 62.4 3e-10
cpv:cgd6_4560 myosin'myosin' 60.1 2e-09
tpv:TP01_0774 myosin 58.2 7e-09
tgo:TGME49_030980 myosin head motor domain-containing protein ... 57.8 7e-09
ath:AT4G33200 XI-I; XI-I; motor/ protein binding 57.0 1e-08
dre:562199 myosin-IXa-like 57.0 1e-08
xla:100158453 myh1; myosin, heavy chain 1, skeletal muscle, ad... 56.2 2e-08
ath:AT3G58160 XIJ; XIJ; motor 55.1 5e-08
dre:100333817 myosin-IXb-like 55.1 6e-08
ath:AT1G08730 XIC; XIC; motor/ protein binding 54.7 8e-08
ath:AT1G54560 XIE; XIE; motor/ protein binding 54.3 9e-08
sce:YHR023W MYO1; Myo1p; K10352 myosin heavy chain 54.3 1e-07
ath:AT1G17580 MYA1; MYA1 (MYOSIN 1); motor/ protein binding 53.9 1e-07
hsa:22989 MYH15; myosin, heavy chain 15; K10352 myosin heavy c... 53.9 1e-07
ath:AT5G20490 XIK; XIK; motor/ protein binding 53.5 1e-07
hsa:4650 MYO9B, CELIAC4, MYR5; myosin IXB; K10360 myosin IX 53.5 2e-07
ath:AT2G31900 XIF; XIF; motor 53.5 2e-07
cel:T10H10.1 hum-6; Heavy chain, Unconventional Myosin family ... 53.5 2e-07
dre:564573 myo3b; myosin IIIB; K08834 myosin III [EC:2.7.11.1] 53.1 2e-07
ath:AT5G43900 MYA2; MYA2 (ARABIDOPSIS MYOSIN 2); GTP-dependent... 53.1 2e-07
mmu:66196 Myo19, 1110055A02Rik, Myohd1; myosin XIX 53.1 2e-07
ath:AT1G04160 XIB; XIB (MYOSIN XI B); motor 53.1 2e-07
ath:AT1G04600 XIA; XIA (MYOSIN XI A); motor/ protein binding 53.1 2e-07
mmu:17925 Myo9b; myosin IXb; K10360 myosin IX 52.8 2e-07
ath:AT2G33240 XID; XID; motor/ protein binding 52.8 3e-07
dre:100006612 myo9al1; myosin IXa-like 1; K10360 myosin IX 52.8 3e-07
ath:AT4G28710 XIH; XIH; motor 52.8 3e-07
cel:F20G4.3 nmy-2; Non-muscle MYosin family member (nmy-2) 52.8 3e-07
xla:380409 myo5a, MGC53270; myosin VA (heavy chain 12, myoxin)... 52.8 3e-07
hsa:64005 MYO1G, HA2, MGC142104, MHAG; myosin IG; K10356 myosin I 52.4 3e-07
cel:F36D4.3 hum-2; Heavy chain, Unconventional Myosin family m... 52.4 3e-07
xla:734396 myo19, MGC115095; myosin XIX 52.4 4e-07
hsa:4649 MYO9A, FLJ11061, FLJ13244, MGC71859; myosin IXA; K103... 52.4 4e-07
dre:402814 myo3a; myosin IIIA (EC:2.7.11.1); K08834 myosin III... 52.4 4e-07
mmu:17922 Myo7b; myosin VIIB; K10359 myosin VII 52.4 4e-07
dre:560956 myo9b, fb53f01, si:dkeyp-66d1.8, wu:fb53f01, wu:fc4... 52.4 4e-07
hsa:4648 MYO7B, DKFZp686A08248; myosin VIIB; K10359 myosin VII 52.0 4e-07
mmu:667663 Myo3a, 9030416P08Rik; myosin IIIA (EC:2.7.11.1); K0... 52.0 4e-07
mmu:270163 Myo9a, 4732465J09Rik, C130068I12Rik, C230003M11; my... 52.0 5e-07
ath:AT4G27370 VIIIB; VIIIB; motor 52.0 5e-07
hsa:140469 MYO3B; myosin IIIB (EC:2.7.11.1); K08834 myosin III... 52.0 5e-07
dre:445473 myo6a; myosin VIa; K10358 myosin VI 52.0 5e-07
dre:556606 myo1ha, myo1h, si:dkey-189p24.1; myosin IHa; K10356... 52.0 5e-07
mmu:667772 Myh15, EG667772; myosin, heavy chain 15; K10352 myo... 51.6 5e-07
hsa:55930 MYO5C, MGC74969; myosin VC; K10357 myosin V 51.6 5e-07
sce:YAL029C MYO4, FUN22, SHE1; Myo4p 51.6 6e-07
mmu:329421 Myo3b, A430065P19Rik; myosin IIIB (EC:2.7.11.1); K0... 51.6 6e-07
sce:YOR326W MYO2, CDC66; Myo2p 51.6 7e-07
> tgo:TGME49_078870 myosin F (TgMyoF) protein (EC:2.7.11.1 1.14.15.6)
Length=1953
Score = 64.7 bits (156), Expect = 7e-11, Method: Composition-based stats.
Identities = 29/49 (59%), Positives = 37/49 (75%), Gaps = 0/49 (0%)
Query 30 GKEEEKNKKEEEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
G +E ++ +LF GVL+IFGFECF+ NS EQLCIN+TNE LQ +FN
Sbjct 500 GYLKEVQSADDLLLFCGVLDIFGFECFQFNSFEQLCINFTNERLQNFFN 548
> pfa:MAL13P1.148 myosin; K01553 myosin ATPase [EC:3.6.4.1]
Length=2160
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 28/36 (77%), Positives = 31/36 (86%), Gaps = 0/36 (0%)
Query 43 LFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
LF GVL+IFGFE F NS EQLCINYTNECLQ++FN
Sbjct 503 LFCGVLDIFGFESFPVNSFEQLCINYTNECLQQFFN 538
> bbo:BBOV_IV011050 23.m05995; WD40 repeat myosin-like protein
(EC:3.6.4.1)
Length=1651
Score = 62.4 bits (150), Expect = 3e-10, Method: Composition-based stats.
Identities = 24/33 (72%), Positives = 29/33 (87%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
G+L+IFGFECF+ NS EQLCIN+TNE LQ +FN
Sbjct 453 GILDIFGFECFQHNSFEQLCINFTNETLQNFFN 485
> cpv:cgd6_4560 myosin'myosin'
Length=1924
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 28/47 (59%), Positives = 36/47 (76%), Gaps = 3/47 (6%)
Query 32 EEEKNKKEEEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
+EK+KK+ L+ G+L+IFGFECF NS EQLCIN+ NE LQ+ FN
Sbjct 561 HQEKDKKD---LYCGILDIFGFECFPNNSFEQLCINFANERLQQIFN 604
> tpv:TP01_0774 myosin
Length=1103
Score = 58.2 bits (139), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 24/33 (72%), Positives = 29/33 (87%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
G+L+IFGFECF+ NS EQLCIN+TNE LQ +FN
Sbjct 514 GILDIFGFECFQLNSFEQLCINFTNETLQNFFN 546
> tgo:TGME49_030980 myosin head motor domain-containing protein
TgMyo I (EC:3.2.1.52 3.6.3.28)
Length=1821
Score = 57.8 bits (138), Expect = 7e-09, Method: Composition-based stats.
Identities = 25/42 (59%), Positives = 32/42 (76%), Gaps = 0/42 (0%)
Query 37 KKEEEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
K+ + LF G+L+I+GFE F NS EQLCINY NE LQ++FN
Sbjct 634 KERDSRLFIGLLDIYGFEVFAANSFEQLCINYANEKLQQHFN 675
> ath:AT4G33200 XI-I; XI-I; motor/ protein binding
Length=1522
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 23/33 (69%), Positives = 28/33 (84%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+I+GFECFK NS EQ CIN+ NE LQ++FN
Sbjct 435 GVLDIYGFECFKNNSFEQFCINFANEKLQQHFN 467
> dre:562199 myosin-IXa-like
Length=1997
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 26/41 (63%), Positives = 30/41 (73%), Gaps = 0/41 (0%)
Query 38 KEEEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
K EIL GVL+IFGFE ++ NS EQ CIN+ NE LQ YFN
Sbjct 206 KLAEILSIGVLDIFGFEDYENNSFEQFCINFANETLQHYFN 246
> xla:100158453 myh1; myosin, heavy chain 1, skeletal muscle,
adult; K10352 myosin heavy chain
Length=1942
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 25/35 (71%), Positives = 29/35 (82%), Gaps = 0/35 (0%)
Query 44 FWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
F GVL+I GFE F NSLEQLCIN+TNE LQ++FN
Sbjct 459 FIGVLDIAGFEIFDFNSLEQLCINFTNEKLQQFFN 493
> ath:AT3G58160 XIJ; XIJ; motor
Length=1242
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 24/33 (72%), Positives = 28/33 (84%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+I+GFE FK NS EQ CINYTNE LQ++FN
Sbjct 429 GVLDIYGFESFKTNSFEQFCINYTNEKLQQHFN 461
> dre:100333817 myosin-IXb-like
Length=709
Score = 55.1 bits (131), Expect = 6e-08, Method: Composition-based stats.
Identities = 29/47 (61%), Positives = 32/47 (68%), Gaps = 4/47 (8%)
Query 36 NKKEEE----ILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
NKK+ E L GVL+IFGFE F+ NS EQ CINY NE LQ YFN
Sbjct 502 NKKDMEESVSCLSIGVLDIFGFEDFETNSFEQFCINYANEQLQYYFN 548
> ath:AT1G08730 XIC; XIC; motor/ protein binding
Length=1538
Score = 54.7 bits (130), Expect = 8e-08, Method: Composition-based stats.
Identities = 23/33 (69%), Positives = 28/33 (84%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+I+GFE FK NS EQ CIN+TNE LQ++FN
Sbjct 439 GVLDIYGFESFKTNSFEQFCINFTNEKLQQHFN 471
> ath:AT1G54560 XIE; XIE; motor/ protein binding
Length=1529
Score = 54.3 bits (129), Expect = 9e-08, Method: Composition-based stats.
Identities = 23/33 (69%), Positives = 28/33 (84%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+I+GFE FK NS EQ CIN+TNE LQ++FN
Sbjct 434 GVLDIYGFESFKTNSFEQFCINFTNEKLQQHFN 466
> sce:YHR023W MYO1; Myo1p; K10352 myosin heavy chain
Length=1928
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 23/35 (65%), Positives = 29/35 (82%), Gaps = 0/35 (0%)
Query 44 FWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
+ G+L+I GFE F+ NS EQLCINYTNE LQ++FN
Sbjct 459 YIGLLDIAGFEIFENNSFEQLCINYTNEKLQQFFN 493
> ath:AT1G17580 MYA1; MYA1 (MYOSIN 1); motor/ protein binding
Length=1520
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 23/33 (69%), Positives = 28/33 (84%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+I+GFE FK NS EQ CIN+TNE LQ++FN
Sbjct 428 GVLDIYGFESFKCNSFEQFCINFTNEKLQQHFN 460
> hsa:22989 MYH15; myosin, heavy chain 15; K10352 myosin heavy
chain
Length=1946
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 23/35 (65%), Positives = 29/35 (82%), Gaps = 0/35 (0%)
Query 44 FWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
F G+L+I GFE + NSLEQLCIN+TNE LQ++FN
Sbjct 468 FIGILDITGFEILEYNSLEQLCINFTNEKLQQFFN 502
> ath:AT5G20490 XIK; XIK; motor/ protein binding
Length=1465
Score = 53.5 bits (127), Expect = 1e-07, Method: Composition-based stats.
Identities = 23/33 (69%), Positives = 28/33 (84%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+I+GFE FK NS EQ CIN+TNE LQ++FN
Sbjct 365 GVLDIYGFESFKINSFEQFCINFTNEKLQQHFN 397
> hsa:4650 MYO9B, CELIAC4, MYR5; myosin IXB; K10360 myosin IX
Length=2022
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 29/47 (61%), Positives = 32/47 (68%), Gaps = 4/47 (8%)
Query 36 NKKEEE----ILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
NKK+ E L GVL+IFGFE F+ NS EQ CINY NE LQ YFN
Sbjct 503 NKKDVEEAVSCLSIGVLDIFGFEDFERNSFEQFCINYANEQLQYYFN 549
> ath:AT2G31900 XIF; XIF; motor
Length=1556
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 24/33 (72%), Positives = 28/33 (84%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+I+GFE FK NS EQLCIN TNE LQ++FN
Sbjct 431 GVLDIYGFESFKINSFEQLCINLTNEKLQQHFN 463
> cel:T10H10.1 hum-6; Heavy chain, Unconventional Myosin family
member (hum-6); K10359 myosin VII
Length=2098
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 21/32 (65%), Positives = 27/32 (84%), Gaps = 0/32 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYF 77
G+L+IFGFE F+ NS EQLCIN+ NE LQ++F
Sbjct 427 GILDIFGFENFESNSFEQLCINFANETLQQFF 458
> dre:564573 myo3b; myosin IIIB; K08834 myosin III [EC:2.7.11.1]
Length=949
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 23/33 (69%), Positives = 27/33 (81%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
G+L+IFGFE FK+NS EQLCIN NE +Q YFN
Sbjct 526 GILDIFGFENFKKNSFEQLCINIANEQIQFYFN 558
> ath:AT5G43900 MYA2; MYA2 (ARABIDOPSIS MYOSIN 2); GTP-dependent
protein binding / Rab GTPase binding / motor
Length=1505
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 23/33 (69%), Positives = 27/33 (81%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+I+GFE FK NS EQ CIN TNE LQ++FN
Sbjct 431 GVLDIYGFESFKTNSFEQFCINLTNEKLQQHFN 463
> mmu:66196 Myo19, 1110055A02Rik, Myohd1; myosin XIX
Length=963
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 22/34 (64%), Positives = 28/34 (82%), Gaps = 0/34 (0%)
Query 44 FWGVLNIFGFECFKENSLEQLCINYTNECLQEYF 77
F G+L+++GFE F NSLEQLCINY NE LQ++F
Sbjct 409 FIGLLDVYGFESFPNNSLEQLCINYANEKLQQHF 442
> ath:AT1G04160 XIB; XIB (MYOSIN XI B); motor
Length=1500
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 23/33 (69%), Positives = 27/33 (81%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+I+GFE FK NS EQ CIN TNE LQ++FN
Sbjct 432 GVLDIYGFESFKTNSFEQFCINLTNEKLQQHFN 464
> ath:AT1G04600 XIA; XIA (MYOSIN XI A); motor/ protein binding
Length=1730
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 23/33 (69%), Positives = 27/33 (81%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+I+GFE FK NS EQ CIN TNE LQ++FN
Sbjct 430 GVLDIYGFESFKTNSFEQFCINLTNEKLQQHFN 462
> mmu:17925 Myo9b; myosin IXb; K10360 myosin IX
Length=2128
Score = 52.8 bits (125), Expect = 2e-07, Method: Composition-based stats.
Identities = 28/46 (60%), Positives = 31/46 (67%), Gaps = 4/46 (8%)
Query 36 NKKEEE----ILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYF 77
NKK+ E L GVL+IFGFE F+ NS EQ CINY NE LQ YF
Sbjct 503 NKKDMEEAVSCLSIGVLDIFGFEDFERNSFEQFCINYANEQLQYYF 548
> ath:AT2G33240 XID; XID; motor/ protein binding
Length=1770
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 23/33 (69%), Positives = 27/33 (81%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+I+GFE FK NS EQ CIN TNE LQ++FN
Sbjct 447 GVLDIYGFESFKTNSFEQFCINLTNEKLQQHFN 479
> dre:100006612 myo9al1; myosin IXa-like 1; K10360 myosin IX
Length=2530
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 24/42 (57%), Positives = 31/42 (73%), Gaps = 0/42 (0%)
Query 37 KKEEEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
++ +IL GVL+IFGFE ++ NS EQ CIN+ NE LQ YFN
Sbjct 525 EESAKILSIGVLDIFGFEDYENNSFEQFCINFANERLQHYFN 566
> ath:AT4G28710 XIH; XIH; motor
Length=1516
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 23/33 (69%), Positives = 27/33 (81%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+I+GFE FK NS EQ CIN TNE LQ++FN
Sbjct 430 GVLDIYGFESFKTNSFEQFCINLTNEKLQQHFN 462
> cel:F20G4.3 nmy-2; Non-muscle MYosin family member (nmy-2)
Length=2003
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 24/41 (58%), Positives = 32/41 (78%), Gaps = 0/41 (0%)
Query 38 KEEEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
+++ + F G+L+I GFE F+ NS EQLCINYTNE LQ+ FN
Sbjct 461 RQQSVSFIGILDIAGFEIFETNSFEQLCINYTNEKLQQLFN 501
> xla:380409 myo5a, MGC53270; myosin VA (heavy chain 12, myoxin);
K10357 myosin V
Length=594
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 23/35 (65%), Positives = 27/35 (77%), Gaps = 0/35 (0%)
Query 44 FWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
F GVL+I+GFE F+ NS EQ CINY NE LQ+ FN
Sbjct 434 FIGVLDIYGFETFEINSFEQFCINYANEKLQQQFN 468
> hsa:64005 MYO1G, HA2, MGC142104, MHAG; myosin IG; K10356 myosin
I
Length=1018
Score = 52.4 bits (124), Expect = 3e-07, Method: Composition-based stats.
Identities = 24/52 (46%), Positives = 35/52 (67%), Gaps = 3/52 (5%)
Query 26 VKKKGKEEEKNKKEEEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYF 77
++ +G++ ++ K+ I GVL+I+GFE F NS EQ CINY NE LQ+ F
Sbjct 375 MEPRGRDPRRDGKDTVI---GVLDIYGFEVFPVNSFEQFCINYCNEKLQQLF 423
> cel:F36D4.3 hum-2; Heavy chain, Unconventional Myosin family
member (hum-2); K10357 myosin V
Length=1839
Score = 52.4 bits (124), Expect = 3e-07, Method: Composition-based stats.
Identities = 26/54 (48%), Positives = 34/54 (62%), Gaps = 0/54 (0%)
Query 25 GVKKKGKEEEKNKKEEEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
+ +K K + N+K+ F GVL+I+GFE F NS EQ INY NE LQ+ FN
Sbjct 469 ALNEKDKLDGTNQKKRPDRFIGVLDIYGFETFDVNSFEQFSINYANEKLQQQFN 522
> xla:734396 myo19, MGC115095; myosin XIX
Length=971
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 22/34 (64%), Positives = 29/34 (85%), Gaps = 0/34 (0%)
Query 44 FWGVLNIFGFECFKENSLEQLCINYTNECLQEYF 77
F G+L+++GFE F EN+LEQLCINY NE LQ++F
Sbjct 420 FIGLLDVYGFESFPENNLEQLCINYANEKLQQHF 453
> hsa:4649 MYO9A, FLJ11061, FLJ13244, MGC71859; myosin IXA; K10360
myosin IX
Length=2548
Score = 52.4 bits (124), Expect = 4e-07, Method: Composition-based stats.
Identities = 27/48 (56%), Positives = 32/48 (66%), Gaps = 3/48 (6%)
Query 31 KEEEKNKKEEEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
K+ E N K I GVL+IFGFE ++ NS EQ CIN+ NE LQ YFN
Sbjct 522 KDLEHNTKTLSI---GVLDIFGFEDYENNSFEQFCINFANERLQHYFN 566
> dre:402814 myo3a; myosin IIIA (EC:2.7.11.1); K08834 myosin III
[EC:2.7.11.1]
Length=1775
Score = 52.4 bits (124), Expect = 4e-07, Method: Composition-based stats.
Identities = 25/48 (52%), Positives = 35/48 (72%), Gaps = 0/48 (0%)
Query 31 KEEEKNKKEEEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
K + ++ ++++ L G+L+IFGFE FK NS EQLCIN NE +Q YFN
Sbjct 711 KPDNQSGEDDKGLNIGILDIFGFENFKRNSFEQLCINIANEQIQFYFN 758
> mmu:17922 Myo7b; myosin VIIB; K10359 myosin VII
Length=2113
Score = 52.4 bits (124), Expect = 4e-07, Method: Composition-based stats.
Identities = 21/32 (65%), Positives = 27/32 (84%), Gaps = 0/32 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYF 77
G+L+IFGFE F+ NS EQLCIN+ NE LQ++F
Sbjct 434 GLLDIFGFENFQNNSFEQLCINFANEHLQQFF 465
> dre:560956 myo9b, fb53f01, si:dkeyp-66d1.8, wu:fb53f01, wu:fc47c07;
myosin IXb; K10360 myosin IX
Length=1937
Score = 52.4 bits (124), Expect = 4e-07, Method: Composition-based stats.
Identities = 24/33 (72%), Positives = 25/33 (75%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
GVL+IFGFE FK NS EQ CINY NE LQ Y N
Sbjct 514 GVLDIFGFEDFKTNSFEQFCINYANEQLQYYCN 546
> hsa:4648 MYO7B, DKFZp686A08248; myosin VIIB; K10359 myosin VII
Length=2116
Score = 52.0 bits (123), Expect = 4e-07, Method: Composition-based stats.
Identities = 21/32 (65%), Positives = 27/32 (84%), Gaps = 0/32 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYF 77
G+L+IFGFE F+ NS EQLCIN+ NE LQ++F
Sbjct 434 GLLDIFGFENFENNSFEQLCINFANEHLQQFF 465
> mmu:667663 Myo3a, 9030416P08Rik; myosin IIIA (EC:2.7.11.1);
K08834 myosin III [EC:2.7.11.1]
Length=1621
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 25/39 (64%), Positives = 29/39 (74%), Gaps = 0/39 (0%)
Query 40 EEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
+E L G+L+IFGFE FK NS EQLCIN NE +Q YFN
Sbjct 715 DEELNIGILDIFGFENFKRNSFEQLCINIANEQIQYYFN 753
> mmu:270163 Myo9a, 4732465J09Rik, C130068I12Rik, C230003M11;
myosin IXa; K10360 myosin IX
Length=2631
Score = 52.0 bits (123), Expect = 5e-07, Method: Composition-based stats.
Identities = 23/42 (54%), Positives = 31/42 (73%), Gaps = 0/42 (0%)
Query 37 KKEEEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
+++ + L GVL+IFGFE ++ NS EQ CIN+ NE LQ YFN
Sbjct 525 EQDTKTLSIGVLDIFGFEDYENNSFEQFCINFANERLQHYFN 566
> ath:AT4G27370 VIIIB; VIIIB; motor
Length=1126
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 21/33 (63%), Positives = 27/33 (81%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
+L+I+GFE FK+NS EQ CINY NE LQ++FN
Sbjct 517 SILDIYGFESFKDNSFEQFCINYANERLQQHFN 549
> hsa:140469 MYO3B; myosin IIIB (EC:2.7.11.1); K08834 myosin III
[EC:2.7.11.1]
Length=1314
Score = 52.0 bits (123), Expect = 5e-07, Method: Composition-based stats.
Identities = 22/33 (66%), Positives = 26/33 (78%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
G+L+IFGFE F+ NS EQLCIN NE +Q YFN
Sbjct 719 GILDIFGFENFQRNSFEQLCINIANEQIQYYFN 751
> dre:445473 myo6a; myosin VIa; K10358 myosin VI
Length=1292
Score = 52.0 bits (123), Expect = 5e-07, Method: Composition-based stats.
Identities = 23/35 (65%), Positives = 27/35 (77%), Gaps = 0/35 (0%)
Query 44 FWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
F GVL+I GFE F+ NS EQ CINY NE LQ++FN
Sbjct 451 FIGVLDIAGFEYFEHNSFEQFCINYCNEKLQQFFN 485
> dre:556606 myo1ha, myo1h, si:dkey-189p24.1; myosin IHa; K10356
myosin I
Length=1164
Score = 52.0 bits (123), Expect = 5e-07, Method: Composition-based stats.
Identities = 24/46 (52%), Positives = 29/46 (63%), Gaps = 0/46 (0%)
Query 32 EEEKNKKEEEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYF 77
E +NK E G+L+I+GFE F NS EQ CINY NE LQ+ F
Sbjct 501 ESLENKDSERKTVIGLLDIYGFEVFTANSFEQFCINYCNEKLQQLF 546
> mmu:667772 Myh15, EG667772; myosin, heavy chain 15; K10352 myosin
heavy chain
Length=1925
Score = 51.6 bits (122), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 22/35 (62%), Positives = 27/35 (77%), Gaps = 0/35 (0%)
Query 44 FWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
F G+L+ GFE NSLEQLCIN+TNE LQ++FN
Sbjct 447 FVGILDTTGFEILDYNSLEQLCINFTNEKLQQFFN 481
> hsa:55930 MYO5C, MGC74969; myosin VC; K10357 myosin V
Length=1742
Score = 51.6 bits (122), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 23/35 (65%), Positives = 26/35 (74%), Gaps = 0/35 (0%)
Query 44 FWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
F GVL+I+GFE F NS EQ CINY NE LQ+ FN
Sbjct 430 FIGVLDIYGFETFDVNSFEQFCINYANEKLQQQFN 464
> sce:YAL029C MYO4, FUN22, SHE1; Myo4p
Length=1471
Score = 51.6 bits (122), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 24/56 (42%), Positives = 37/56 (66%), Gaps = 0/56 (0%)
Query 23 LGGVKKKGKEEEKNKKEEEILFWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
+ + K + E ++++ F G+L+I+GFE F++NS EQ CINY NE LQ+ FN
Sbjct 423 VDNINKTLYDPELDQQDHVFSFIGILDIYGFEHFEKNSFEQFCINYANEKLQQEFN 478
> mmu:329421 Myo3b, A430065P19Rik; myosin IIIB (EC:2.7.11.1);
K08834 myosin III [EC:2.7.11.1]
Length=1333
Score = 51.6 bits (122), Expect = 6e-07, Method: Composition-based stats.
Identities = 22/33 (66%), Positives = 26/33 (78%), Gaps = 0/33 (0%)
Query 46 GVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
G+L+IFGFE F+ NS EQLCIN NE +Q YFN
Sbjct 735 GILDIFGFEDFQRNSFEQLCINIANEQIQYYFN 767
> sce:YOR326W MYO2, CDC66; Myo2p
Length=1574
Score = 51.6 bits (122), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 23/35 (65%), Positives = 28/35 (80%), Gaps = 0/35 (0%)
Query 44 FWGVLNIFGFECFKENSLEQLCINYTNECLQEYFN 78
F GVL+I+GFE F++NS EQ CINY NE LQ+ FN
Sbjct 443 FIGVLDIYGFEHFEKNSFEQFCINYANEKLQQEFN 477
Lambda K H
0.311 0.135 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2067704464
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40