bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2280_orf1
Length=137
Score E
Sequences producing significant alignments: (Bits) Value
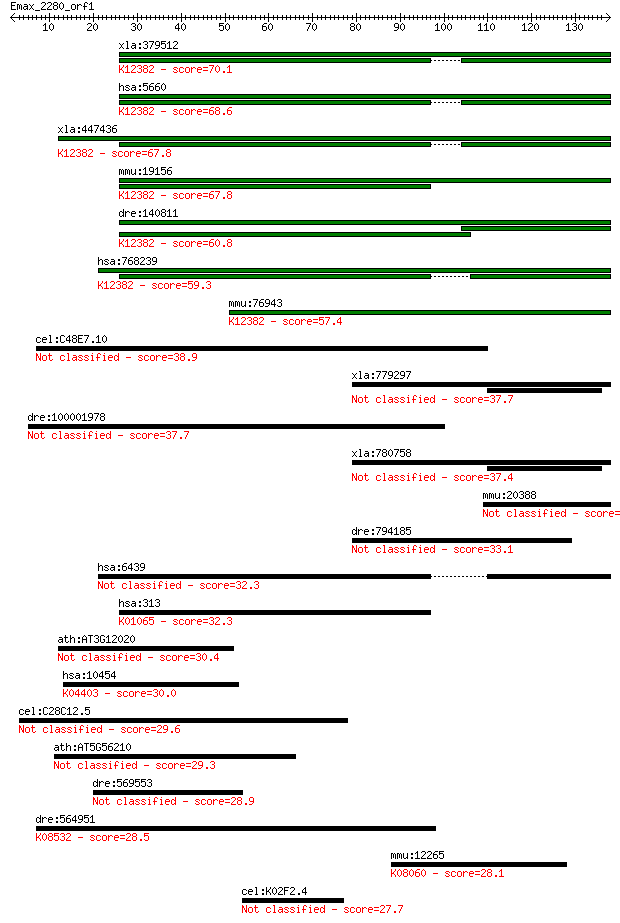
xla:379512 hypothetical protein MGC64541; K12382 saposin 70.1 2e-12
hsa:5660 PSAP, FLJ00245, GLBA, MGC110993, SAP1; prosaposin; K1... 68.6 6e-12
xla:447436 psap; prosaposin (variant Gaucher disease and varia... 67.8 9e-12
mmu:19156 Psap, AI037048, SGP-1; prosaposin; K12382 saposin 67.8 1e-11
dre:140811 psap, cb759, wu:fa14a06, wu:fb36e02, wu:fb58g07; pr... 60.8 1e-09
hsa:768239 PSAPL1, FLJ40379; prosaposin-like 1 (gene/pseudogen... 59.3 4e-09
mmu:76943 Psapl1, 2310020A21Rik; prosaposin-like 1; K12382 sap... 57.4 1e-08
cel:C48E7.10 spp-15; SaPosin-like Protein family member (spp-15) 38.9 0.005
xla:779297 sftpb, MGC154673, SP-B, xSP-B; surfactant, pulmonar... 37.7 0.009
dre:100001978 wu:fd19f07; si:ch211-283h6.6 37.7 0.011
xla:780758 surfactant protein B 37.4 0.012
mmu:20388 Sftpb, AI562151, SF-B, SP-B, Sftp-3, Sftp3; surfacta... 35.0 0.067
dre:794185 hypothetical LOC794185 33.1 0.24
hsa:6439 SFTPB, PSP-B, SFTB3, SFTP3, SMDP1, SP-B; surfactant p... 32.3 0.42
hsa:313 AOAH; acyloxyacyl hydrolase (neutrophil) (EC:3.1.1.77)... 32.3 0.46
ath:AT3G12020 kinesin motor protein-related 30.4 1.8
hsa:10454 TAB1, 3'-Tab1, MAP3K7IP1, MGC57664; TGF-beta activat... 30.0 2.1
cel:C28C12.5 spp-8; SaPosin-like Protein family member (spp-8) 29.6 3.2
ath:AT5G56210 WIP2; WIP2 (WPP-domain Interacting Protein 2); p... 29.3 3.7
dre:569553 syne1a, si:ch211-225l4.1, si:ch211-87a14.2, wu:fc76... 28.9 5.0
dre:564951 roraa, RORalpha-B, gb:dq017624, rora2; RAR-related ... 28.5 6.4
mmu:12265 Ciita, C2ta, EG669998, Gm9475, MGC130299; class II t... 28.1 7.9
cel:K02F2.4 ulp-5; Ubiquitin-Like Protease family member (ulp-5) 27.7 9.9
> xla:379512 hypothetical protein MGC64541; K12382 saposin
Length=512
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 34/112 (30%), Positives = 55/112 (49%), Gaps = 2/112 (1%)
Query 26 CEMCQNIVNNLQQVQSHDLELDQLTENSCSKLPKAHRTECGLMMKAFAPYFFDMMNHQDN 85
C M V+ L + + + + C+ LP + + EC ++K + P F ++ +
Sbjct 401 CRMIMRYVDELLEKNATESRIKDFLNRICNFLPDSMQNECSALIKEYEPLFIQLLLEALD 460
Query 86 AKEICKSIDICMTPGHVHLLGGHKCTFGPSYWCHTVDHAHACNAATYCRQKV 137
IC + +C G LLG KC +GPSYWC V+ A CNA +C++ V
Sbjct 461 PSFICLKLHLCQ--GETVLLGTEKCMWGPSYWCKDVETAANCNALEHCQRHV 510
Score = 37.0 bits (84), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 17/34 (50%), Gaps = 0/34 (0%)
Query 104 LLGGHKCTFGPSYWCHTVDHAHACNAATYCRQKV 137
L G +C GP WC V A C A +C+Q V
Sbjct 19 LFGTEQCAKGPEVWCENVRTASQCGAVKHCQQSV 52
Score = 28.9 bits (63), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 13/74 (17%), Positives = 37/74 (50%), Gaps = 3/74 (4%)
Query 26 CEMCQNIVNNLQQVQSHDLE---LDQLTENSCSKLPKAHRTECGLMMKAFAPYFFDMMNH 82
CE+C+ +++ ++++ ++ + Q E C LP + +C ++ + +++
Sbjct 303 CEVCELMISQIEKLLDNNRTRENIKQSLEKVCKLLPSQYTQKCEDIIDEYCDPLIELLEQ 362
Query 83 QDNAKEICKSIDIC 96
+ N + IC ++ C
Sbjct 363 EANPEVICTTLGYC 376
> hsa:5660 PSAP, FLJ00245, GLBA, MGC110993, SAP1; prosaposin;
K12382 saposin
Length=527
Score = 68.6 bits (166), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 35/115 (30%), Positives = 56/115 (48%), Gaps = 4/115 (3%)
Query 26 CEMCQNIVNNLQ---QVQSHDLELDQLTENSCSKLPKAHRTECGLMMKAFAPYFFDMMNH 82
CE+C+ +V L + S E+ E CS LP ++ +C + + P +++
Sbjct 412 CEVCKKLVGYLDRNLEKNSTKQEILAALEKGCSFLPDPYQKQCDQFVAEYEPVLIEILVE 471
Query 83 QDNAKEICKSIDICMTPGHVHLLGGHKCTFGPSYWCHTVDHAHACNAATYCRQKV 137
+ +C I C + H LLG KC +GPSYWC + A CNA +C++ V
Sbjct 472 VMDPSFVCLKIGACPS-AHKPLLGTEKCIWGPSYWCQNTETAAQCNAVEHCKRHV 525
Score = 33.5 bits (75), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 18/34 (52%), Gaps = 0/34 (0%)
Query 104 LLGGHKCTFGPSYWCHTVDHAHACNAATYCRQKV 137
+LG +CT G + WC V A C A +C Q V
Sbjct 19 VLGLKECTRGSAVWCQNVKTASDCGAVKHCLQTV 52
Score = 30.0 bits (66), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 15/74 (20%), Positives = 36/74 (48%), Gaps = 3/74 (4%)
Query 26 CEMCQNIVNNLQQV---QSHDLELDQLTENSCSKLPKAHRTECGLMMKAFAPYFFDMMNH 82
CE+C+ +V + ++ + E+ + CSKLPK+ EC ++ + ++
Sbjct 318 CEVCEFLVKEVTKLIDNNKTEKEILDAFDKMCSKLPKSLSEECQEVVDTYGSSILSILLE 377
Query 83 QDNAKEICKSIDIC 96
+ + + +C + +C
Sbjct 378 EVSPELVCSMLHLC 391
> xla:447436 psap; prosaposin (variant Gaucher disease and variant
metachromatic leukodystrophy); K12382 saposin
Length=518
Score = 67.8 bits (164), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 39/134 (29%), Positives = 64/134 (47%), Gaps = 10/134 (7%)
Query 12 SDNKLIVLNNAEKR--------CEMCQNIVNNLQQVQSHDLELDQLTENSCSKLPKAHRT 63
S N IV +AEK C+M V+ L + + ++ + C+ LP + +
Sbjct 385 SKNLKIVKISAEKAAAGDYCAVCKMLMRYVDELLEKNATEIRIKAFLGRICNFLPDSMQN 444
Query 64 ECGLMMKAFAPYFFDMMNHQDNAKEICKSIDICMTPGHVHLLGGHKCTFGPSYWCHTVDH 123
EC ++ + P F ++ + IC +++C LLG KC +GPSYWC ++
Sbjct 445 ECSALVNEYEPLFIQLLLEALDPSFICIKVNLCQ--NKKVLLGTEKCMWGPSYWCKDMET 502
Query 124 AHACNAATYCRQKV 137
A CNA +CR+ V
Sbjct 503 AANCNALEHCRRHV 516
Score = 38.5 bits (88), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 18/34 (52%), Gaps = 0/34 (0%)
Query 104 LLGGHKCTFGPSYWCHTVDHAHACNAATYCRQKV 137
L G +C GP WC TV A C A +C+Q V
Sbjct 19 LFGTEQCAKGPEVWCETVRTASQCGAVKHCQQNV 52
Score = 33.5 bits (75), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 15/74 (20%), Positives = 39/74 (52%), Gaps = 3/74 (4%)
Query 26 CEMCQNIVNNLQQVQSHDLELDQL---TENSCSKLPKAHRTECGLMMKAFAPYFFDMMNH 82
CE+C+ +V+ L+++ ++ + + E C LP + +C M++ ++ +++
Sbjct 309 CEVCELMVSQLEKLLDNNRTRENIKHGLEKVCKLLPSQYTQKCEDMIEEYSDALIELLEQ 368
Query 83 QDNAKEICKSIDIC 96
+ N + IC ++ C
Sbjct 369 EANPQAICTALGYC 382
> mmu:19156 Psap, AI037048, SGP-1; prosaposin; K12382 saposin
Length=554
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 34/115 (29%), Positives = 59/115 (51%), Gaps = 4/115 (3%)
Query 26 CEMCQNIVNNLQ---QVQSHDLELDQLTENSCSKLPKAHRTECGLMMKAFAPYFFDMMNH 82
CE+C+ +V L+ + S E+ E CS LP ++ +C + + P +++
Sbjct 439 CEVCKKLVLYLEHNLEKNSTKEEILAALEKGCSFLPDPYQKQCDDFVAEYEPLLLEILVE 498
Query 83 QDNAKEICKSIDICMTPGHVHLLGGHKCTFGPSYWCHTVDHAHACNAATYCRQKV 137
+ +C I +C + + LLG KC +GPSYWC ++ A CNA +C++ V
Sbjct 499 VMDPGFVCSKIGVCPS-AYKLLLGTEKCVWGPSYWCQNMETAARCNAVDHCKRHV 552
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 20/74 (27%), Positives = 38/74 (51%), Gaps = 3/74 (4%)
Query 26 CEMCQNIVNNLQQVQSHDLELDQLTE---NSCSKLPKAHRTECGLMMKAFAPYFFDMMNH 82
C+ CQ ++N ++ ++ + L + N+C+ LP RT+C ++ F P D+ H
Sbjct 314 CQTCQFVMNKFSELIVNNATEELLVKGLSNACALLPDPARTKCQEVVGTFGPSLLDIFIH 373
Query 83 QDNAKEICKSIDIC 96
+ N +C I +C
Sbjct 374 EVNPSSLCGVIGLC 387
> dre:140811 psap, cb759, wu:fa14a06, wu:fb36e02, wu:fb58g07;
prosaposin; K12382 saposin
Length=522
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 33/113 (29%), Positives = 58/113 (51%), Gaps = 3/113 (2%)
Query 26 CEMCQNIVNNLQQVQSHDLELDQLTENSCSKLPKAHRTECGLMMKAFAPYFFDMMNHQDN 85
C+M V+ + + + E+++ CS LP A + EC +++ + P ++ +
Sbjct 410 CKMAVRYVDGILEQNATQSEIEEAVLKVCSFLPYAVKDECNQLIEQYEPLLVQLLLQTLD 469
Query 86 AKEICKSIDICMTPGHVH-LLGGHKCTFGPSYWCHTVDHAHACNAATYCRQKV 137
+C + C P V LLG ++C++GP+YWC V A CNA +CR+ V
Sbjct 470 PDFVCMKLGAC--PEAVQRLLGLNQCSWGPAYWCKNVQTAARCNALNHCRRHV 520
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 16/34 (47%), Positives = 19/34 (55%), Gaps = 0/34 (0%)
Query 104 LLGGHKCTFGPSYWCHTVDHAHACNAATYCRQKV 137
LLG +C GP YWC V A C A +C+Q V
Sbjct 17 LLGTEQCARGPPYWCQNVKTASLCGAVQHCQQNV 50
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 18/80 (22%), Positives = 38/80 (47%), Gaps = 0/80 (0%)
Query 26 CEMCQNIVNNLQQVQSHDLELDQLTENSCSKLPKAHRTECGLMMKAFAPYFFDMMNHQDN 85
CE + N+ Q Q+ + E+ Q E C+ LP +C +++ + D++ + +
Sbjct 317 CEYVMKEIENMIQDQTSEAEIVQAVEKVCNILPSTLTAQCKDLIETYGQAIIDLLVQEAD 376
Query 86 AKEICKSIDICMTPGHVHLL 105
K +C + +C HV ++
Sbjct 377 PKTVCSFLALCSGVSHVPVM 396
> hsa:768239 PSAPL1, FLJ40379; prosaposin-like 1 (gene/pseudogene);
K12382 saposin
Length=521
Score = 59.3 bits (142), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 37/124 (29%), Positives = 57/124 (45%), Gaps = 9/124 (7%)
Query 21 NAEKRCEMCQNIVNNLQQVQSHDLE-----LDQLT--ENSCSKLPKAHRTECGLMMKAFA 73
+AE + C N L V SH+LE D L + CS LP + +C + +
Sbjct 388 DAENQGSFC-NGCKRLLTVSSHNLESKSTKRDILVAFKGGCSILPLPYMIQCKHFVTQYE 446
Query 74 PYFFDMMNHQDNAKEICKSIDICMTPGHVHLLGGHKCTFGPSYWCHTVDHAHACNAATYC 133
P + + + +CK + C P LLG +C GPS+WC + + A CNA +C
Sbjct 447 PVLIESLKDMMDPVAVCKKVGACHGP-RTPLLGTDQCALGPSFWCRSQEAAKLCNAVQHC 505
Query 134 RQKV 137
++ V
Sbjct 506 QKHV 509
Score = 36.2 bits (82), Expect = 0.033, Method: Compositional matrix adjust.
Identities = 19/74 (25%), Positives = 37/74 (50%), Gaps = 4/74 (5%)
Query 26 CEMCQNIVNNLQ---QVQSHDLELDQLTENSCSKLPKAHRTECGLMMKAFAPYFFDMMNH 82
CE+C N+V L S +L + E CS +P + EC +++ ++P ++
Sbjct 294 CEVCMNVVQKLDHWLMSNSSELMITHALERVCSVMPASITKECIILVDTYSPSLVQLVA- 352
Query 83 QDNAKEICKSIDIC 96
+ +++CK I +C
Sbjct 353 KITPEKVCKFIRLC 366
Score = 29.3 bits (64), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 11/32 (34%), Positives = 15/32 (46%), Gaps = 0/32 (0%)
Query 106 GGHKCTFGPSYWCHTVDHAHACNAATYCRQKV 137
G +C G + WC + A C A YC+ V
Sbjct 22 GPQECAKGSTVWCQDLQTAARCGAVGYCQGAV 53
> mmu:76943 Psapl1, 2310020A21Rik; prosaposin-like 1; K12382 saposin
Length=525
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 25/87 (28%), Positives = 42/87 (48%), Gaps = 1/87 (1%)
Query 51 ENSCSKLPKAHRTECGLMMKAFAPYFFDMMNHQDNAKEICKSIDICMTPGHVHLLGGHKC 110
+ C LP + +C + + P + + N ++CK + C P LLG +C
Sbjct 428 KGGCRILPLPYVMQCNRFVAEYEPVLIESLKFMMNPTDLCKKMGACHGP-KTPLLGTDQC 486
Query 111 TFGPSYWCHTVDHAHACNAATYCRQKV 137
GPS+WC + + A CNA +C++ V
Sbjct 487 VMGPSFWCKSPEAAEMCNALEHCQRLV 513
> cel:C48E7.10 spp-15; SaPosin-like Protein family member (spp-15)
Length=118
Score = 38.9 bits (89), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 24/104 (23%), Positives = 46/104 (44%), Gaps = 1/104 (0%)
Query 7 TKPNISDNKLIVLNNAEKRCEMCQNIVNNLQQVQSHDLELDQLTENSCSK-LPKAHRTEC 65
T N+ K + + ++ C +C N+V L QV H +++ + C + +P C
Sbjct 15 TVSNVEGAKKMHSDTSKPLCGLCVNVVKQLDQVLEHGGDIEAAVDKFCKEDVPSFMVDMC 74
Query 66 GLMMKAFAPYFFDMMNHQDNAKEICKSIDICMTPGHVHLLGGHK 109
+++ Y + + + A +IC I +C TP + L K
Sbjct 75 EKVIEKNLEYIINKLKDHEEADKICTDILLCRTPKQYYFLETQK 118
> xla:779297 sftpb, MGC154673, SP-B, xSP-B; surfactant, pulmonary-associated
protein B
Length=393
Score = 37.7 bits (86), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 18/60 (30%), Positives = 31/60 (51%), Gaps = 3/60 (5%)
Query 79 MMNHQDNAKEICKSIDICMTPGHVHLLGGHK-CTFGPSYWCHTVDHAHACNAATYCRQKV 137
+++ Q + + C+++ C P + H CT GPSYWC ++ A C A ++C V
Sbjct 334 LLHKQWDHRMTCQALGACPVPANAA--AQHSGCTVGPSYWCQNLETAKECGAVSHCLTHV 391
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 10/26 (38%), Positives = 14/26 (53%), Gaps = 0/26 (0%)
Query 110 CTFGPSYWCHTVDHAHACNAATYCRQ 135
C GP +WC + A C A +C+Q
Sbjct 28 CALGPEFWCQDLMTAAQCGAVDHCKQ 53
> dre:100001978 wu:fd19f07; si:ch211-283h6.6
Length=317
Score = 37.7 bits (86), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 19/95 (20%), Positives = 42/95 (44%), Gaps = 0/95 (0%)
Query 5 VCTKPNISDNKLIVLNNAEKRCEMCQNIVNNLQQVQSHDLELDQLTENSCSKLPKAHRTE 64
+ TKP+ + +N C + V +L + + +++L E C LP+ +
Sbjct 134 IYTKPSRGTGPEVRINPICNFCLLFIKTVESLLPKEKTEAAIEELLEKICGYLPEHYEDT 193
Query 65 CGLMMKAFAPYFFDMMNHQDNAKEICKSIDICMTP 99
C +K +A +++ + IC ++ +C+ P
Sbjct 194 CNTFVKTYAKQLIELLLYSMPPHAICTALGLCLLP 228
> xla:780758 surfactant protein B
Length=393
Score = 37.4 bits (85), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 18/60 (30%), Positives = 31/60 (51%), Gaps = 3/60 (5%)
Query 79 MMNHQDNAKEICKSIDICMTPGHVHLLGGHK-CTFGPSYWCHTVDHAHACNAATYCRQKV 137
+++ Q + + C+++ C P + H CT GPSYWC ++ A C A ++C V
Sbjct 334 LLHKQWDHRMTCQALGACPVPANAA--AQHSGCTVGPSYWCQNLETAKDCGAVSHCLTHV 391
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 10/26 (38%), Positives = 14/26 (53%), Gaps = 0/26 (0%)
Query 110 CTFGPSYWCHTVDHAHACNAATYCRQ 135
C GP +WC + A C A +C+Q
Sbjct 28 CALGPEFWCQDLMTAAQCGAVDHCKQ 53
> mmu:20388 Sftpb, AI562151, SF-B, SP-B, Sftp-3, Sftp3; surfactant
associated protein B
Length=377
Score = 35.0 bits (79), Expect = 0.067, Method: Compositional matrix adjust.
Identities = 12/29 (41%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 109 KCTFGPSYWCHTVDHAHACNAATYCRQKV 137
+C GP +WC +++HA C A +C Q+V
Sbjct 30 ECAQGPQFWCQSLEHAVQCRALGHCLQEV 58
> dre:794185 hypothetical LOC794185
Length=215
Score = 33.1 bits (74), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 13/50 (26%), Positives = 23/50 (46%), Gaps = 0/50 (0%)
Query 79 MMNHQDNAKEICKSIDICMTPGHVHLLGGHKCTFGPSYWCHTVDHAHACN 128
++ Q A + CKS ++C + ++ C+ GP Y C + A C
Sbjct 161 FLSKQQGALDTCKSANLCPSDDKESVITADPCSLGPKYSCRDLQTAVECG 210
> hsa:6439 SFTPB, PSP-B, SFTB3, SFTP3, SMDP1, SP-B; surfactant
protein B
Length=393
Score = 32.3 bits (72), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 11/28 (39%), Positives = 17/28 (60%), Gaps = 0/28 (0%)
Query 110 CTFGPSYWCHTVDHAHACNAATYCRQKV 137
C GP +WC +++ A C A +C Q+V
Sbjct 44 CAQGPEFWCQSLEQALQCRALGHCLQEV 71
Score = 31.2 bits (69), Expect = 0.93, Method: Compositional matrix adjust.
Identities = 19/80 (23%), Positives = 38/80 (47%), Gaps = 4/80 (5%)
Query 21 NAEKRCEMCQNIVNNLQQVQSHDLELD---QLTENSCSKLP-KAHRTECGLMMKAFAPYF 76
A+ C+ C++IV+ L ++ + D + E C+ LP K +C ++ + P
Sbjct 76 GADDLCQECEDIVHILNKMAKEAIFQDTMRKFLEQECNVLPLKLLMPQCNQVLDDYFPLV 135
Query 77 FDMMNHQDNAKEICKSIDIC 96
D +Q ++ IC + +C
Sbjct 136 IDYFQNQTDSNGICMHLGLC 155
> hsa:313 AOAH; acyloxyacyl hydrolase (neutrophil) (EC:3.1.1.77);
K01065 acyloxyacyl hydrolase [EC:3.1.1.77]
Length=688
Score = 32.3 bits (72), Expect = 0.46, Method: Composition-based stats.
Identities = 19/74 (25%), Positives = 37/74 (50%), Gaps = 3/74 (4%)
Query 26 CEMCQNIVNNLQQV-QSHDLELDQLTENSCSKLPKAH--RTECGLMMKAFAPYFFDMMNH 82
C C +V+ ++Q+ Q H+ + E CS LP+ +T C L++ F +++
Sbjct 41 CVGCVLVVSVIEQLAQVHNSTVQASMERLCSYLPEKLFLKTTCYLVIDKFGSDIIKLLSA 100
Query 83 QDNAKEICKSIDIC 96
NA +C +++ C
Sbjct 101 DMNADVVCHTLEFC 114
> ath:AT3G12020 kinesin motor protein-related
Length=1030
Score = 30.4 bits (67), Expect = 1.8, Method: Composition-based stats.
Identities = 18/42 (42%), Positives = 24/42 (57%), Gaps = 2/42 (4%)
Query 12 SDNKLI--VLNNAEKRCEMCQNIVNNLQQVQSHDLELDQLTE 51
+DN++I LN CE+ Q V NL+Q S LEL Q T+
Sbjct 801 ADNRIIQQTLNEKTCECEVLQEEVANLKQQLSEALELAQGTK 842
> hsa:10454 TAB1, 3'-Tab1, MAP3K7IP1, MGC57664; TGF-beta activated
kinase 1/MAP3K7 binding protein 1; K04403 TAK1-binding
protein 1
Length=504
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 22/40 (55%), Gaps = 3/40 (7%)
Query 13 DNKLIVLNNAEKRCEMCQNIVNNLQQVQSHDLELDQLTEN 52
+NKL V N R +C++ V+ LQ Q L +D TEN
Sbjct 175 NNKLYVANVGTNRALLCKSTVDGLQVTQ---LNVDHTTEN 211
> cel:C28C12.5 spp-8; SaPosin-like Protein family member (spp-8)
Length=402
Score = 29.6 bits (65), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 36/80 (45%), Gaps = 8/80 (10%)
Query 3 IHVCTKPNISDNKLIVLNNAEK---RCEMCQNIVNNLQQVQS--HDLELDQLTENSCSKL 57
+H C K +N L+ + +EK CE C+ + + Q Q H +D L N C KL
Sbjct 309 MHSCEKK---ENALVEMAMSEKVMLGCENCKAVEHFFAQNQEALHSHAVDGLYSNVCQKL 365
Query 58 PKAHRTECGLMMKAFAPYFF 77
P A T C + + FF
Sbjct 366 PTALGTMCEASIIRLSRKFF 385
> ath:AT5G56210 WIP2; WIP2 (WPP-domain Interacting Protein 2);
protein heterodimerization/ protein homodimerization
Length=509
Score = 29.3 bits (64), Expect = 3.7, Method: Composition-based stats.
Identities = 15/55 (27%), Positives = 27/55 (49%), Gaps = 0/55 (0%)
Query 11 ISDNKLIVLNNAEKRCEMCQNIVNNLQQVQSHDLELDQLTENSCSKLPKAHRTEC 65
I + K + L+ +K +N+ N Q +Q+H +E+ ++ E C K T C
Sbjct 430 IEEEKTLGLSKLDKAETKAENLKNQAQDLQNHCVEITEIQEVECLKKRAFKTTRC 484
> dre:569553 syne1a, si:ch211-225l4.1, si:ch211-87a14.2, wu:fc76h10;
spectrin repeat containing, nuclear envelope 1a
Length=8650
Score = 28.9 bits (63), Expect = 5.0, Method: Composition-based stats.
Identities = 13/34 (38%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 20 NNAEKRCEMCQNIVNNLQQVQSHDLELDQLTENS 53
N E+ C + NI +Q++QSH +L+Q +E S
Sbjct 1617 NLEERWCSLSTNIPLRIQELQSHLAQLEQFSEVS 1650
> dre:564951 roraa, RORalpha-B, gb:dq017624, rora2; RAR-related
orphan receptor A, paralog a; K08532 RAR-related orphan receptor
alpha
Length=468
Score = 28.5 bits (62), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 23/93 (24%), Positives = 39/93 (41%), Gaps = 3/93 (3%)
Query 7 TKPNISDNKL--IVLNNAEKRCEMCQNIVNNLQQVQSHDLELDQLTENSCSKLPKAHRTE 64
T P +S +L + N ++ E CQ + LQQ+ L + EN SK +
Sbjct 209 TSPTVSMAELEHLAQNISKSHMETCQYLREELQQMTWQAF-LQEEVENYQSKPREVMWQL 267
Query 65 CGLMMKAFAPYFFDMMNHQDNAKEICKSIDICM 97
C + + Y + H D E+C++ I +
Sbjct 268 CAIKITEAIQYVVEFAKHIDGFMELCQNDQIVL 300
> mmu:12265 Ciita, C2ta, EG669998, Gm9475, MGC130299; class II
transactivator; K08060 class II, major histocompatibility complex,
transactivator
Length=1078
Score = 28.1 bits (61), Expect = 7.9, Method: Composition-based stats.
Identities = 16/43 (37%), Positives = 21/43 (48%), Gaps = 5/43 (11%)
Query 88 EICKSIDICMTPGH---VHLLGGHKCTFGPSYWCHTVDHAHAC 127
E+ + + C PG V +LG K G S+W TV H AC
Sbjct 347 EVLQVVSDCRRPGETQVVAVLG--KAGQGKSHWARTVSHTWAC 387
> cel:K02F2.4 ulp-5; Ubiquitin-Like Protease family member (ulp-5)
Length=311
Score = 27.7 bits (60), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 12/24 (50%), Positives = 16/24 (66%), Gaps = 1/24 (4%)
Query 54 CSKLPK-AHRTECGLMMKAFAPYF 76
C KLP+ + +CG+ M AFA YF
Sbjct 242 CQKLPQQKNSVDCGIFMMAFAEYF 265
Lambda K H
0.323 0.133 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2428006156
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40